Metabolic Responses of Poplar to Apripona germari (Hope) as Revealed by Metabolite Profiling
Abstract
:1. Introduction
2. Results
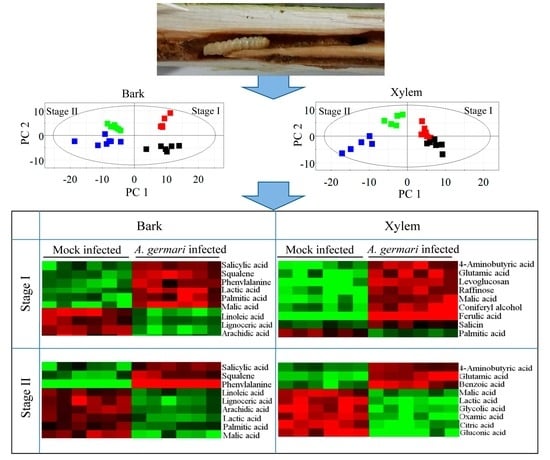
2.1. Metabolic Profiles of Bark and Xylem Tissues in A. germari and Mock Infected Trees
2.2. Metabolic Alteration in Bark and Xylem Tissues Following Infection of A. germari
3. Discussion
4. Materials and Methods
4.1. Insect Bioassays
4.2. Metabolite Extraction and Derivatization
4.3. Analysis of Metabolites by GC–TOF/MS
4.4. Data Processing and Statistical Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hu, J.J.; Wang, L.J.; Yan, D.H.; Lu, M.Z. Research and application of transgenic poplar in China. In Challenges and Opportunities for the World's Forests in the 21st Century; Fenning, T., Ed.; Springer: Dordrecht, The Netherlands, 2014; pp. 567–584. [Google Scholar]
- Liu, H.M.; Sun, X.Y.; Wang, X.G. Advances in the research on Apriona germari Hope. For. Pest Dis. 2002, 21, 30–33. [Google Scholar]
- Mithöfer, A.; Boland, W. Plant defense against herbivores: Chemical aspects. Annu. Rev. Plant Biol. 2012, 63, 431–450. [Google Scholar] [CrossRef] [PubMed]
- Karban, R.; Baldwin, I.T. Induced Responses to Herbivory; University of Chicago Press: Chicago, IL, USA, 1997. [Google Scholar]
- Agrawal, A.A. Induced responses to herbivory and increased plant performance. Science 1998, 279, 1201–1202. [Google Scholar] [CrossRef] [PubMed]
- Young, B.; Wagner, D.; Doak, P.; Clausen, T. Induction of phenolic glycosides by quaking aspen (Populus tremuloides) leaves in relation to extra floral nectaries and epidermal leaf mining. J. Chem. Ecol. 2010, 36, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Peters, D.J.; Constabel, C.P. Molecular analysis of herbivore-induced condensed tannin synthesis: Cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J. 2002, 32, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.J. Mapping of QTLs for Concentration of Phenolic Glycosides and Insect Resistance in Populus deltoides. Ph.D. Thesis, Chinese Academy of Forestry Press, Beijing, China, 2000. [Google Scholar]
- Schauer, N.; Fernie, A.R. Plant metabolomics: Towards biological function and mechanism. Trends Plant Sci. 2006, 11, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Hao, F.; Hu, J.; Zhang, W.; Wan, L.L.; Zhu, L.L.; Tang, H.R.; He, G.C. Revealing different systems responses to brown planthopper infestation for pest susceptible and resistant rice plants with the combined metabonomic and gene-expression analysis. J. Proteome Res. 2010, 9, 6774–6785. [Google Scholar] [CrossRef] [PubMed]
- Marti, G.; Erb, M.; Boccard, J.; Glauser, G.; Doyen, G.R.; Villard, N.; Brobert, C.A.M.; Turlings, T.C.J.; Rudaz, S.; Wolfender, J.L. Metabolomics reveals herbivore-induced metabolites of resistance and susceptibility in maize leaves and roots. Plant Cell Environ. 2013, 36, 621–639. [Google Scholar] [CrossRef] [PubMed]
- Agut, B.; Gamir, J.; Jaques, J.A.; Flors, V. Tetranychus urticae-triggered responses promote genotype-dependent conspecific repellence or attractiveness in citrus. New Phytol. 2015, 207, 790–804. [Google Scholar] [CrossRef] [PubMed]
- Constabel, C.P.; Barbehenn, R. Defensive roles of polyphenol oxidase in plants. In Induced Plant Resistance to Herbivory; Springer: Dordrecht, The Netherlands, 2008; pp. 253–270. [Google Scholar]
- Barbehenn, R.V.; Constabel, C.P. Tannins in plant–herbivore interactions. Phytochemistry 2011, 72, 1551–1565. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, T. Biosynthesis of lignin. In Plant Carbohydrates II; Springer: Berlin/Heidelberg, Germay, 1981; pp. 194–224. [Google Scholar]
- Brodeur-Campbell, S.E.; Vucetich, J.A.; Richter, D.L.; Waite, T.A.; Rosemier, J.N.; Tsai, C.-J. Insect herbivory on low-lignin transgenic aspen. Environ. Entomol. 2006, 35, 1696–1701. [Google Scholar] [CrossRef]
- Schroeder, F.C.; del Campo, M.L.; Grant, J.B.; Weibel, D.B.; Smedley, S.R.; Bolton, K.L.; Meinwald, J.; Eisner, T. Pinoresinol: A lignol of plant origin serving for defense in a caterpillar. Proc. Natl. Acad. Sci. USA 2006, 103, 15497–15501. [Google Scholar] [CrossRef] [PubMed]
- Felton, G.W.; Donato, K.; Delvecchio, R.J.; Duffey, S.S. Activation of plant foliar oxidases by insect feeding reduces nutritive quality of foliage for noctuid herbibores. J. Chem. Ecol. 1989, 15, 2667–2694. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, P.C.; Anderson, J.C.; Blaney, W.M.; Simmonds, M.S.J. Developmental inhibition of Spodoptera litura (Fab) larvae by a novel caffeoyl quinic acid form the wild groundnut, Arachis paraguariensis (Chod et Hassl). J. Chem. Ecol. 1993, 19, 2917–2933. [Google Scholar] [CrossRef] [PubMed]
- Plaxton, W.C. The organization and regulation of plant glycolysis. Annu. Rev. Plant Biol. 1996, 47, 185–214. [Google Scholar] [CrossRef] [PubMed]
- Sweetlove, L.J.; Beard, K.F.; Nunes-Nesi, A.; Fernie, A.R.; Ratcliffe, R.G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010, 15, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef] [PubMed]
- McConn, M.; Creelman, R.A.; Bell, E.; Mullet, J.E. Jasmonate is essential for insect defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 5473–5477. [Google Scholar] [CrossRef] [PubMed]
- Loake, G.; Grant, M. Salicylic acid in plant defence—The players and protagonists. Curr. Opin. Plant Biol. 2007, 10, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Bari, R.; Achard, P.; Lisón, P.; Nemri, A.; Harberd, N.P.; Jones, J.D. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 2008, 18, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Walling, L.L. Avoiding effective defenses: Strategies employed by phloem-feeding insects. Plant Physiol. 2008, 146, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Tzin, V.; Fernandez-Pozo, N.; Richter, A.; Schmelz, E.A.; Schoettner, M.; Schäfer, M.; Mori, N. Dynamic maize responses to aphid feeding are revealed by a time series of transcriptomic and metabolomic assays. Plant Physiol. 2015, 169, 1727–1743. [Google Scholar] [CrossRef] [PubMed]
- Hilfiker, O.; Groux, R.; Bruessow, F.; Kiefer, K.; Zeier, J.; Reymond, P. Insect eggs induce a systemic acquired resistance in Arabidopsis. Plant J. 2014, 80, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Jander, G.; de Vos, M. Non-protein amino acids in plant defense against insect herbivores: Representative cases and opportunities for further functional analysis. Phytochemistry 2011, 72, 1531–1537. [Google Scholar] [CrossRef] [PubMed]
- Ramputh, A.I.; Bown, A.W. Rapid γ-aminobutyric acid synthesis and the inhibition of the growth and development of oblique-banded leaf-roller larvae. Plant Physiol. 1996, 111, 1349–1352. [Google Scholar] [PubMed]
- Kinnersley, A.M.; Turano, F.J. Gamma aminobutyric acid (GABA) and plant responses to stress. Crit. Rev. Plant Sci. 2000, 19, 479–509. [Google Scholar] [CrossRef]
- Bouche, N.; Fromm, H. GABA in plants: Just a metabolite? Trends Plant Sci. 2004, 9, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Scholz, S.S.; Reichelt, M.; Mekonnen, D.W.; Ludewig, F.; Mithöfer, A. Insect herbivory-elicited GABA accumulation in plants is a wound-induced, direct, systemic, and jasmonate-independent defense response. Front. Plant Sci. 2015, 6, 1128. [Google Scholar] [CrossRef] [PubMed]
- Weckwerth, W.; Wenzel, K.; Fiehn, O. Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics 2004, 4, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.J.; Wang, L.J.; Wang, Q.H.; Wang, Y.Z.; Zhang, Y.A. Metabolic profiling of somatic tissues from Monochamus alternatus (Coleoptera: Cerambycidae) reveals effects of irradiation on metabolism. Int. J. Mol. Sci. 2014, 15, 10806–10820. [Google Scholar] [CrossRef] [PubMed]
- Mev software v4.9. Available online: http://source forge.net/projects/mev-tm4 (accessed on 4 April 2013).
- Xia, J.; Sinelnikov, I.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef] [PubMed]





© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Qu, L.; Zhang, L.; Hu, J.; Tang, F.; Lu, M. Metabolic Responses of Poplar to Apripona germari (Hope) as Revealed by Metabolite Profiling. Int. J. Mol. Sci. 2016, 17, 923. https://doi.org/10.3390/ijms17060923
Wang L, Qu L, Zhang L, Hu J, Tang F, Lu M. Metabolic Responses of Poplar to Apripona germari (Hope) as Revealed by Metabolite Profiling. International Journal of Molecular Sciences. 2016; 17(6):923. https://doi.org/10.3390/ijms17060923
Chicago/Turabian StyleWang, Lijuan, Liangjian Qu, Liwei Zhang, Jianjun Hu, Fang Tang, and Mengzhu Lu. 2016. "Metabolic Responses of Poplar to Apripona germari (Hope) as Revealed by Metabolite Profiling" International Journal of Molecular Sciences 17, no. 6: 923. https://doi.org/10.3390/ijms17060923
APA StyleWang, L., Qu, L., Zhang, L., Hu, J., Tang, F., & Lu, M. (2016). Metabolic Responses of Poplar to Apripona germari (Hope) as Revealed by Metabolite Profiling. International Journal of Molecular Sciences, 17(6), 923. https://doi.org/10.3390/ijms17060923