Human Milk Cells Contain Numerous miRNAs that May Change with Milk Removal and Regulate Multiple Physiological Processes
Abstract
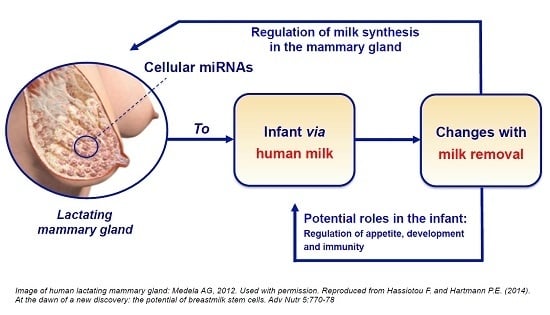
:1. Introduction
2. Results
2.1. Human Milk Cells Contain Numerous Known and Novel miRNAs
2.2. Effects of Feeding on the miRNA Content of Human Milk Cells
2.3. Human Milk Cellular miRNAs Are Regulatory Agents in the Mammary Gland
3. Discussion
4. Materials and Methods
4.1. Ethics Statement and Sample Collection
4.2. Cell Isolation and miRNA Extraction from Human Milk
4.3. Statistical Analysis of Human Milk Composition
4.4. Library Construction, Small RNA Sequencing, and Bioinformatics Analysis
4.5. Differential Expression Analysis
4.6. qPCR Validation
4.7. Target Prediction and Functional Analysis
4.8. Analysis of Pathways, Networks, and miRNA Gene Targets
4.9. Availability of Supporting Data
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hassiotou, F.; Geddes, D. Anatomy of the human mammary gland: Current status of knowledge. Clin. Anat. 2013, 26, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Stevens, E.E.; Patrick, T.E.; Pickler, R. A history of infant feeding. J. Perinat. Educ. 2009, 18, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. MicroRNAs in breastmilk and the lactating breast: Potential immunoprotectors and developmental regulators for the infant and the mother. Int. J. Environ. Res. Publ. Health 2015, 12, 13981–14020. [Google Scholar] [CrossRef] [PubMed]
- Hassiotou, F.; Geddes, D.T. Immune cell-mediated protection of the mammary gland and the infant during breastfeeding. Adv. Nutr. 2015, 6, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Hassiotou, F.; Hartmann, P.E. At the dawn of a new discovery: The potential of breast milk stem cells. Adv. Nutr. 2014, 5, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Hassiotou, F.; Hepworth, A.R.; Metzger, P.; Tat Lai, C.; Trengove, N.; Hartmann, P.E.; Filgueira, L. Maternal and infant infections stimulate a rapid leukocyte response in breastmilk. Clin. Transl. Immunol. 2013, 2, e3. [Google Scholar] [CrossRef] [PubMed]
- Riskin, A.; Almog, M.; Peri, R.; Halasz, K.; Srugo, I.; Kessel, A. Changes in immunomodulatory constituents of human milk in response to active infection in the nursing infant. Pediatr. Res. 2012, 71, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. microRNA as a new immune-regulatory agent in breast milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Munch, E.M.; Harris, R.A.; Mohammad, M.; Benham, A.L.; Pejerrey, S.M.; Showalter, L.; Hu, M.; Shope, C.D.; Maningat, P.D.; Gunaratne, P.H.; et al. Transcriptome profiling of microRNA by Next-Gen deep sequencing reveals known and novel miRNA species in the lipid fraction of human breast milk. PLoS ONE 2013, 8, e50564. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Tat-Lai, C.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci. Rep. 2016, 6, 20680. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Hepworth, A.R.; Lefevre, C.; Hartmann, P.E.; Geddes, D.T.; Hassiotou, F. Human milk microRNA and total RNA differ depending on milk fractionation. J. Cell. Biochem. 2015, 116, 2397–2407. [Google Scholar] [CrossRef] [PubMed]
- Lasser, C.; Alikhani, V.S.; Ekstrom, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjostrand, M.; Gabrielsson, S.; Lotvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filen, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Baier, S.R.; Nguyen, C.; Xie, F.; Wood, J.R.; Zempleni, J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J. Nutr. 2014, 144, 1495–1500. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Sang, X.; Hong, Z. Beyond nutrients: Food-derived microRNAs provide cross-kingdom regulation. Bioessays 2012, 34, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wiggins, B.E.; Lawrence, C.; Petrick, J.; Ivashuta, S.; Heck, G. Analysis of plant-derived miRNAs in animal small RNA datasets. BMC Genom. 2012, 13, 381. [Google Scholar] [CrossRef] [PubMed]
- Arntz, O.J.; Pieters, B.C.; Oliveira, M.C.; Broeren, M.G.; Bennink, M.B.; De Vries, M.; van Lent, P.L.; Koenders, M.I.; Van Den Berg, W.B.; Van Der Kraan, P.M.; et al. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol. Nutr. Food Res. 2015, 59, 1701–1712. [Google Scholar] [CrossRef] [PubMed]
- Wolf, T.; Baier, S.R.; Zempleni, J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma caco-2 cells and rat small intestinal IEC-6 cells. J. Nutr. 2015, 145, 2201–2206. [Google Scholar] [CrossRef] [PubMed]
- Vaucheret, H.; Chupeau, Y. Ingested plant miRNAs regulate gene expression in animals. Cell Res. 2012, 22, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, X.; Liu, J.; Dong, L.; Chen, Q.; Liu, J.; Kong, H.; Zhang, Q.; Qi, X.; Hou, D.; et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015, 25, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, B.; Zhang, Y.; Petrick, J.S.; Heck, G.; Ivashuta, S.; Marshall, W.S. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol. 2013, 31, 965–967. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Farmer, L.M.; Agyekum, A.A.; Hirschi, K.D. Detection of dietary plant-based small RNAs in animals. Cell Res. 2015, 25, 517–520. [Google Scholar] [CrossRef] [PubMed]
- Fallingborg, J. Intraluminal pH of the human gastrointestinal tract. Dan. Med. Bull. 1999, 46, 183–196. [Google Scholar] [PubMed]
- Chen, X.; Gao, C.; Li, H.; Huang, L.; Sun, Q.; Dong, Y.; Tian, C.; Gao, S.; Dong, H.; Guan, D.; et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010, 20, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Time-dependent expression profiles of microRNAs and mRNAs in rat milk whey. PLoS ONE 2014, 9, e88843. [Google Scholar] [CrossRef] [PubMed]
- Modepalli, V.; Kumar, A.; Hinds, L.A.; Sharp, J.A.; Nicholas, K.R.; Lefevre, C. Differential temporal expression of milk miRNA during the lactation cycle of the marsupial tammar wallaby (Macropus eugenii). BMC Genom. 2014, 15, 1012. [Google Scholar] [CrossRef] [PubMed]
- Hassiotou, F.; Hepworth, A.R.; Williams, T.M.; Twigger, A.J.; Perrella, S.; Lai, C.T.; Filgueira, L.; Geddes, D.T.; Hartmann, P.E. Breastmilk cell and fat contents respond similarly to removal of breastmilk by the infant. PLoS ONE 2013, 8, e78232. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Hepworth, A.R.; Prime, D.K.; Lai, C.T.; Trengove, N.J.; Hartmann, P.E. Variation in fat, lactose, and protein composition in breast milk over 24 hours: Associations with infant feeding patterns. J. Hum. Lactation off. J. Int. Lactation Consult. Assoc. 2013, 29, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Hassiotou, F.; Mobley, A.; Geddes, D.; Hartmann, P.; Wilkie, T. Breastmilk imparts the mother’s stem cells to the infant. FASEB J. 2015, 29, 876–878. [Google Scholar]
- Hassiotou, F.; Geddes, D.T.; Hartmann, P.E. Cells in human milk: State of the science. J. Hum. Lactation Off. J. Int. Lactation Consult. Assoc. 2013, 29, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Mar-Aguilar, F.; Luna-Aguirre, C.M.; Moreno-Rocha, J.C.; Araiza-Chavez, J.; Trevino, V.; Rodriguez-Padilla, C.; Resendez-Perez, D. Differential expression of miR-21, miR-125b and miR-191 in breast cancer tissue. Asia Pac. J. Clin. Oncol. 2013, 9, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, N.; Ahmad, H.M.; Molparia, B.; Kulshreshtha, R. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis 2013, 34, 1889–1899. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Dong, J.; Wang, L.E.; Ma, H.; Liu, J.; Zhao, Y.; Tang, J.; Chen, X.; Dai, J.; Wei, Q.; et al. Serum microRNA profiling and breast cancer risk: The use of miR-484/191 as endogenous controls. Carcinogenesis 2012, 33, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Marchand, A.; Proust, C.; Morange, P.E.; Lompre, A.M.; Tregouet, D.A. miR-421 and miR-30c inhibit SERPINE 1 gene expression in human endothelial cells. PLoS ONE 2012, 7, e44532. [Google Scholar] [CrossRef] [PubMed]
- Isolauri, E.; Sutas, Y.; Kankaanpaa, P.; Arvilommi, H.; Salminen, S. Probiotics: Effects on immunity. Am. J. Clin. Nutr. 2001, 73, 444S–450S. [Google Scholar] [CrossRef] [PubMed]
- Maningat, P.D.; Sen, P.; Sunehag, A.L.; Hadsell, D.L.; Haymond, M.W. Regulation of gene expression in human mammary epithelium: Effect of breast pumping. J. Endocrinol. 2007, 195, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Zempleni, J.; Baier, S.R.; Hirschi, K. Diet-responsive MicroRNAs are likely exogenous. J. Biol. Chem. 2015, 290, 25197. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA. 2012, 109, E2110–E2116. [Google Scholar] [CrossRef] [PubMed]
- Bryniarski, K.; Ptak, W.; Martin, E.; Nazimek, K.; Szczepanik, M.; Sanak, M.; Askenase, P.W. Free extracellular miRNA functionally targets cells by transfecting exosomes from their companion cells. PLoS ONE 2015, 10, e0122991. [Google Scholar]
- Brini, M.; Carafoli, E. Calcium pumps in health and disease. Physiol. Rev. 2009, 89, 1341–1378. [Google Scholar] [CrossRef] [PubMed]
- Hallberg, L.; Rossander-Hulten, L.; Brune, M.; Gleerup, A. Bioavailability in man of iron in human milk and cow’s milk in relation to their calcium contents. Pediatr. Res. 1992, 31, 524–527. [Google Scholar] [CrossRef] [PubMed]
- Kanyshkova, T.G.; Babina, S.E.; Semenov, D.V.; Isaeva, N.; Vlassov, A.V.; Neustroev, K.N.; Kul’minskaya, A.A.; Buneva, V.N.; Nevinsky, G.A. Multiple enzymic activities of human milk lactoferrin. Eur. J. Biochem. 2003, 270, 3353–3361. [Google Scholar] [CrossRef] [PubMed]
- Walker, W.A. The dynamic effects of breastfeeding on intestinal development and host defense. Adv. Exp. Med. Biol. 2004, 554, 155–170. [Google Scholar] [PubMed]
- Dewey, K.G.; Heinig, M.J.; Nommsen-Rivers, L.A. Differences in morbidity between breast-fed and formula-fed infants. J. Pediatr. 1995, 126, 696–702. [Google Scholar] [CrossRef]
- Hanson, L.A.; Winberg, J. Breast milk and defence against infection in the newborn. Archives Dis. Child. 1972, 47, 845–848. [Google Scholar] [CrossRef]
- Howie, P.W.; Forsyth, J.S.; Ogston, S.A.; Clark, A.; Florey, C.D. Protective effect of breast feeding against infection. BMJ 1990, 300, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Duijts, L.; Jaddoe, V.W.; Hofman, A.; Moll, H.A. Prolonged and exclusive breastfeeding reduces the risk of infectious diseases in infancy. Pediatrics 2010, 126, e18–e25. [Google Scholar] [CrossRef] [PubMed]
- Bahl, R.; Frost, C.; Kirkwood, B.R.; Edmond, K.; Martines, J.; Bhandari, N.; Arthur, P. Infant feeding patterns and risks of death and hospitalization in the first half of infancy: Multicenter cohort study. Bull. World Health Organ. 2005, 83, 418–426. [Google Scholar] [PubMed]
- Ford, K.; Labbok, M. Breast-feeding and child health in the United States. J. Biosoc. Sci. 1993, 25, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Cesar, J.A.; Victora, C.G.; Barros, F.C.; Santos, I.S.; Flores, J.A. Impact of breast feeding on admission for pneumonia during postneonatal period in Brazil: Nested case-control study. BMJ 1999, 318, 1316–1320. [Google Scholar] [CrossRef] [PubMed]
- Innis, S.M. Human milk and formula fatty acids. J. Pediatr. 1992, 120, S56–S61. [Google Scholar] [CrossRef]
- Mitoulas, L.R.; Gurrin, L.C.; Doherty, D.A.; Sherriff, J.L.; Hartmann, P.E. Infant intake of fatty acids from human milk over the first year of lactation. Br. J. Nutr. 2003, 90, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Morera Pons, S.; Castellote Bargallo, A.I.; Lopez Sabater, M.C. Analysis of human milk triacylglycerols by high-performance liquid chromatography with light-scattering detection. J. Chromatogr. A 1998, 823, 475–482. [Google Scholar] [CrossRef]
- Hale, T.W.; Hartmann, P.E. Hale & Hartmann’s Textbook of Human Lactation, 1st ed.; Hale Pub.: Amarillo, TX, USA, 2007; p. 661. [Google Scholar]
- Takeuchi, K.; Reue, K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1195–E1209. [Google Scholar] [CrossRef] [PubMed]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Hambraeus, L. Composition of human milk: Nutritional aspects. Bibl. Nutr. Dieta. 1996, 53, 37–44. [Google Scholar] [PubMed]
- Kunz, C.; Rudloff, S.; Baier, W.; Klein, N.; Strobel, S. Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annu. Rev. Nutr. 2000, 20, 699–722. [Google Scholar] [CrossRef] [PubMed]
- Kulski, J.K.; Hartmann, P.E. Milk insulin GH and TSH: Relationship to changes in milk lactose, glucose and protein during lactogenesis in women. Endocrinol. Exp. 1983, 17, 317–326. [Google Scholar] [PubMed]
- Jensen, R.G. Handbook of Milk Composition; Academic Press: San Diego, CA, USA, 1995; p. 919. [Google Scholar]
- Kelly, P.A.; Bachelot, A.; Kedzia, C.; Hennighausen, L.; Ormandy, C.J.; Kopchick, J.J.; Binart, N. The role of prolactin and growth hormone in mammary gland development. Mol. Cell. Endocrinol. 2002, 197, 127–131. [Google Scholar] [CrossRef]
- Herrington, J.; Carter-Su, C. Signaling pathways activated by the growth hormone receptor. Trends Endocrinol. Metab. 2001, 12, 252–257. [Google Scholar] [CrossRef]
- Wang, Y.D.; Wood, W.I. Amino acids of the human growth hormone receptor that are required for proliferation and Jak-STAT signaling. Mol. Endocrinol. 1995, 9, 303–311. [Google Scholar] [PubMed]
- Nagashima, K.; Itoh, K.; Kuroume, T. Levels of insulin-like growth factor I in full- and preterm human milk in comparison to levels in cow’s milk and in milk formulas. Neonatology 1990, 58, 343–346. [Google Scholar] [CrossRef]
- Buyukkayhan, D.; Tanzer, F.; Erselcan, T.; Cinar, Z.; Yonem, O. Umbilical serum insulin-like growth factor 1 (IGF-1) in newborns: Effects of gestational age, postnatal age, and nutrition. Int. J. Vitam. Nutr. Res. 2003, 73, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Donovan, S.M.; Hintz, R.L.; Rosenfeld, R.G. Insulin-like growth factors I and II and their binding proteins in human milk: Effect of heat treatment on IGF and IGF binding protein stability. J. Pediatr. Gastroenterol. Nutr. 1991, 13, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Strack, A.M.; Sebastian, R.J.; Schwartz, M.W.; Dallman, M.F. Glucocorticoids and insulin: Reciprocal signals for energy balance. Am. J. Physiol. 1995, 268, R142–R149. [Google Scholar] [PubMed]
- Kinouchi, T.; Koizumi, K.; Kuwata, T.; Yajima, T. Milk-borne insulin with trypsin inhibitor in milk induces pancreatic amylase development at the onset of weaning in rats. J. Pediatr. Gastroenterol. Nutr. 2000, 30, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Georgiev, I.P.; Georgieva, T.M.; Pfaffl, M.; Hammon, H.M.; Blum, J.W. Insulin-like growth factor and insulin receptors in intestinal mucosa of neonatal calves. J. Endocrinol. 2003, 176, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Shehadeh, N.; Shamir, R.; Berant, M.; Etzioni, A. Insulin in human milk and the prevention of type 1 diabetes. Pediatr. Diabetes 2001, 2, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Karolina, D.S.; Armugam, A.; Tavintharan, S.; Wong, M.T.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS ONE 2011, 6, e22839. [Google Scholar] [CrossRef]
- Neville, M.C.; McFadden, T.B.; Forsyth, I. Hormonal regulation of mammary differentiation and milk secretion. J. Mammary Gl. Biol. Neoplasia 2002, 7, 49–66. [Google Scholar] [CrossRef]
- Wan, Z.Y.; Tong, H.L.; Li, Q.Z.; Gao, X.J. Influence on cellular signal transduction pathway in dairy cow mammary gland epithelial cells by galactopoietic compound isolated from vaccariae segetalis. Agric. Sci. China 2011, 10, 619–630. [Google Scholar] [CrossRef]
- Zwart, W.; Theodorou, V.; Carroll, J.S. Estrogen receptor-positive breast cancer: A multidisciplinary challenge. Wiley Interdiscip Rev. Syst. Biol. Med. 2011, 3, 216–230. [Google Scholar] [CrossRef] [PubMed]
- O’Day, E.; Lal, A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010, 12, 201. [Google Scholar] [CrossRef] [PubMed]
- Si, M.L.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y.Y. miR-21-mediated tumor growth. Oncogene 2007, 26, 2799–2803. [Google Scholar] [CrossRef] [PubMed]
- Heneghan, H.M.; Miller, N.; Lowery, A.J.; Sweeney, K.J.; Kerin, M.J. MicroRNAs as novel biomarkers for breast cancer. J. Oncol. 2009, 2009, 950201. [Google Scholar] [CrossRef] [PubMed]
- Hassiotou, F.; Beltran, A.; Chetwynd, E.; Stuebe, A.M.; Twigger, A.J.; Metzger, P.; Trengove, N.; Lai, C.T.; Filgueira, L.; Blancafort, P.; et al. Breastmilk is a novel source of stem cells with multilineage differentiation potential. Stem Cells 2012, 30, 2164–2174. [Google Scholar] [CrossRef] [PubMed]
- Team, R. RStudio: Integrated Development for R. Available online: http://www. RStudio. com/ide (accessed on 13 June 2016).
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; R Core Team. Nlme: Linear and Nonlinear Mixed Effects Models. Available online: http://CRAN.R-project.org/package=nlme (accessed on 13 June 2016).
- Sarkar, D. Lattice: Lattice Graphics. Available online: https://stat.ethz.ch/R-manual/R-devel/library/lattice/html/Lattice.html (accessed on 13 June 2016).
- Yi, S.; Gao, Z.X.; Zhao, H.; Zeng, C.; Luo, W.; Chen, B.; Wang, W.M. Identification and characterization of microRNAs involved in growth of blunt snout bream (Megalobrama amblycephala) by Solexa sequencing. BMC Genom. 2013, 14, 754. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Qin, C.; Chen, Z.; Zuo, T.; Yang, X.; Zhou, H.; Xu, M.; Cao, S.; Shen, Y.; Lin, H.; et al. Identification of miRNAs and their target genes in developing maize ears by combined small RNA and degradome sequencing. BMC Genom. 2014, 15, 25. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]







| Sample ID * | Number of Known miRNAs | Number of Known miRNA Reads | Number of Novel miRNAs | Number of Novel miRNA Reads |
|---|---|---|---|---|
| 1_B | 790 | 10,037,707 | 107 | 1507 |
| 1_A | 859 | 9,203,046 | 74 | 1374 |
| 2_B | 563 | 6,483,425 | 209 | 2267 |
| 2_A | 532 | 8,344,307 | 249 | 3667 |
| 3_B | 840 | 10,137,502 | 108 | 1182 |
| 3_A | 876 | 9,424,413 | 103 | 1045 |
| 4_B | 820 | 9,539,244 | 90 | 1138 |
| 4_A | 814 | 9,694,427 | 90 | 1242 |
| 5_B | 750 | 5,658,703 | 35 | 366 |
| 5_A | 823 | 8,164,895 | 53 | 709 |
| 6_B | 833 | 9,378,155 | 140 | 1410 |
| 6_A | 899 | 8,811,523 | 131 | 1224 |
| 7_B | 468 | 6,534,047 | 184 | 1918 |
| 7_A | 662 | 8,058,217 | 157 | 1268 |
| 8_B | 759 | 9,732,524 | 115 | 1240 |
| 8_A | 707 | 7,599,182 | 118 | 1178 |
| 9_B | 750 | 7,744,119 | 143 | 1310 |
| 9_A | 586 | 7,897,409 | 222 | 2009 |
| 10_B | 789 | 10,327,701 | 179 | 1864 |
| 10_A | 780 | 11,415,988 | 330 | 3405 |
| Total: 1467 | 174,186,534 | Total: 1996 | 31,323 |
| Sample Group | Pre-Feed Milk (n = 10) | Post-Feed Milk (n = 10) |
|---|---|---|
| Number of known miRNAs | 1287 | 1308 |
| Total reads (known) | 85,573,127 | 88,613,407 |
| Number of novel miRNAs | 961 (35 *) | 1215 (41 *) |
| Total reads (novel) | 14,202 (5282 *) | 17,121 (6950 *) |
| Number of specific miRNAs | 159 | 180 |
| Mean ± S.D. of the Number of Total Milk cells/mL Milk (Cell Viability %) | Mean ± S.D. of Total miRNA Content (ng/106 cells) | Mean ± S.D. of miRNA Quality (OD 260/280) & RIN | |||
|---|---|---|---|---|---|
| All samples (n = 20) | 1,222,860 ± 767,091 (92.7%) | NanoDrop | 1414 ± 519 | NanoDrop | 2.05 ± 0.05 |
| Bioanalyzer | 1000 ± 438 | Bioanalyzer | 8.67 ± 1.22 | ||
| Pre-feed milk (n = 10) | 1,146,364 ± 843,594 (91.3%) | NanoDrop | 1391 ± 571 | NanoDrop | 2.04 ± 0.06 |
| Bioanalyzer | 996 ± 481 | Bioanalyzer | 8.57 ± 1.69 | ||
| Post-feed milk (n = 10) | 1,299,356 ± 719,432 (93.3%) | NanoDrop | 1438 ± 490 | NanoDrop | 2.05 ± 0.04 |
| Bioanalyzer | 1004 ± 417 | Bioanalyzer | 8.77 ± 0.52 | ||
| Known miRNA | Number of Reads | Number of Samples Detected in (n = 20) | Novel miRNA | Number of Reads | Number of Samples Detected in (n = 20) | |
|---|---|---|---|---|---|---|
| 1 | hsa-let-7f-5p | 25,479,884 | 20 | novel_mir_7 | 3890 | 13 |
| 2 | hsa-miR-181a-5p | 21,601,482 | 20 | novel_mir_299 | 942 | 19 |
| 3 | hsa-miR-148a-3p | 14,925,495 | 20 | novel_mir_367 | 804 | 17 |
| 4 | hsa-miR-22-3p | 12,999,002 | 20 | novel_mir_39 | 789 | 20 |
| 5 | hsa-miR-182-5p | 11,288,364 | 20 | novel_mir_115 | 760 | 17 |
| 6 | hsa-let-7a-5p | 10,343,580 | 20 | novel_mir_476 | 658 | 4 |
| 7 | hsa-miR-375 | 9,145,308 | 20 | novel_mir_90 | 586 | 17 |
| 8 | hsa-miR-141-3p | 8,577,962 | 20 | novel_mir_41 | 341 | 12 |
| 9 | hsa-miR-30a-5p | 7,844,936 | 20 | novel_mir_269 | 322 | 15 |
| 10 | hsa-miR-30d-5p | 6,173,556 | 20 | novel_mir_161 | 295 | 9 |
| 11 | hsa-miR-146b-5p | 4,140,259 | 20 | novel_mir_278 | 247 | 14 |
| 12 | hsa-miR-26a-5p | 3,876,162 | 20 | novel_mir_76 | 222 | 13 |
| 13 | hsa-miR-21-5p | 3,457,523 | 20 | novel_mir_430 | 194 | 14 |
| 14 | hsa-let-7i-5p | 2,264,279 | 20 | novel_mir_144 | 189 | 3 |
| 15 | hsa-miR-92a-3p | 1,918,918 | 20 | novel_mir_456 | 178 | 9 |
| 16 | hsa-miR-27b-3p | 1,849,454 | 20 | novel_mir_202 | 174 | 12 |
| 17 | hsa-miR-181b-5p | 1,594,211 | 20 | novel_mir_159 | 173 | 12 |
| 18 | hsa-miR-423-5p | 1,374,443 | 20 | novel_mir_411 | 169 | 11 |
| 19 | hsa-miR-125a-5p | 1,260,164 | 20 | novel_mir_251 | 156 | 8 |
| 20 | hsa-miR-10a-5p | 1,145,612 | 20 | novel_mir_425 | 154 | 7 |
| Top Function or Disease Involvement | p Value | Number of miRNAs |
|---|---|---|
| Molecular and Cellular Functions | ||
| Cell development | 5.00 × 10−2–6.09 × 10−10 | 24 |
| Cell growth and proliferation | 4.88 × 10−2–6.09 × 10−10 | 26 |
| Cell movement | 4.21 × 10−2–1.17 × 10−6 | 15 |
| Cell death and survival | 4.41 × 10−2–2.45 × 10−5 | 17 |
| Cell cycle | 3.82 × 10−2–3.80 × 10−5 | 9 |
| Physiological System Development and Function | ||
| Organismal development | 4.39 × 10−6–9.21 × 10−10 | 5 |
| Digestive system development and function | 2.54 × 10−6–2.54 × 10−6 | 4 |
| Hepatic system development and function | 2.54 × 10−6–2.54 × 10−6 | 4 |
| Organ development | 1.63 × 10−2–2.54 × 10−6 | 5 |
| Connective tissue development and function | 6.13 × 10−3–3.80 × 10−5 | 4 |
| Diseases and Disorders | ||
| Cancer | 4.41 × 10−2–1.21 × 10−34 | 41 |
| Hematological diseases | 3.01 × 10−2–1.21 × 10−34 | 26 |
| Immunological diseases | 3.01 × 10−2–1.21 × 10−34 | 24 |
| Organismal injury and abnormalities | 4.41 × 10−2–1.21 × 10−34 | 41 |
| Reproductive system diseases | 2.60 × 10−2–2.10 × 10−28 | 31 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human Milk Cells Contain Numerous miRNAs that May Change with Milk Removal and Regulate Multiple Physiological Processes. Int. J. Mol. Sci. 2016, 17, 956. https://doi.org/10.3390/ijms17060956
Alsaweed M, Lai CT, Hartmann PE, Geddes DT, Kakulas F. Human Milk Cells Contain Numerous miRNAs that May Change with Milk Removal and Regulate Multiple Physiological Processes. International Journal of Molecular Sciences. 2016; 17(6):956. https://doi.org/10.3390/ijms17060956
Chicago/Turabian StyleAlsaweed, Mohammed, Ching Tat Lai, Peter E. Hartmann, Donna T. Geddes, and Foteini Kakulas. 2016. "Human Milk Cells Contain Numerous miRNAs that May Change with Milk Removal and Regulate Multiple Physiological Processes" International Journal of Molecular Sciences 17, no. 6: 956. https://doi.org/10.3390/ijms17060956
APA StyleAlsaweed, M., Lai, C. T., Hartmann, P. E., Geddes, D. T., & Kakulas, F. (2016). Human Milk Cells Contain Numerous miRNAs that May Change with Milk Removal and Regulate Multiple Physiological Processes. International Journal of Molecular Sciences, 17(6), 956. https://doi.org/10.3390/ijms17060956