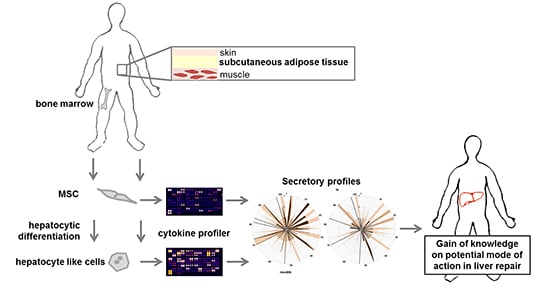
Identification of Pathways in Liver Repair Potentially Targeted by Secretory Proteins from Human Mesenchymal Stem Cells
Abstract
:1. Introduction
2. Results
2.1. Phenotypic Characteristics
2.2. Identification of Hepatotropic Factors Secreted by Mesenchymal Stem Cells (MSC)
2.3. Identification of Pathways and Networks Affected by Factors Secreted by MSC
3. Discussion
3.1. Paracrine Mechanisms in Liver Repair
3.2. Functional Relevance
4. Experimental Section
4.1. Human Material
4.2. Mesenchymal Stem Cell Isolation, Propagation and Hepatocytic Differentiation
4.3. Microscopic Documentation of Morphology
4.4. Flow Cytometry
4.5. Sample Preparation and Proteome Profiler Antibody Array
4.6. Array Analysis and Bioinformatics
4.7. Statistics
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| hbmMSC | Human bone marrow-derived mesenchymal stem cells |
| hsubMSC | Human subcutaneous adipose tissue-derived mesenchymal stem cells |
References
- De Almeida, D.C.; Donizetti-Oliveira, C.; Barbosa-Costa, P.; Origassa, C.S.; Camara, N.O. In search of mechanisms associated with mesenchymal stem cell-based therapies for acute kidney injury. Clin. Biochem. Rev. Aust. Assoc. Clin. Biochem. 2013, 34, 131–144. [Google Scholar]
- Conese, M.; Piro, D.; Carbone, A.; Castellani, S.; di Gioia, S. Hematopoietic and mesenchymal stem cells for the treatment of chronic respiratory diseases: Role of plasticity and heterogeneity. Sci. World J. 2014. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, R.P.; Sturm, M.J. Adult human mesenchymal stromal cells and the treatment of graft versus host disease. Stem Cells Cloning Adv. Appl. 2014, 7, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, G.M.; Gowing, G.; Svendsen, S.; Svendsen, C.N. The past, present and future of stem cell clinical trials for ALS. Exp. Neurol. 2014, 262B, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Fernandez, M.; Rodriguez-Frutos, B.; Ramos-Cejudo, J.; Otero-Ortega, L.; Fuentes, B.; Diez-Tejedor, E. Stem cells for brain repair and recovery after stroke. Expert Opin. Biol. Ther. 2013, 13, 1479–1483. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, S.; Gu, S.; Sang, L.; Dai, C. Mesenchymal stem cells recruit macrophages to alleviate experimental colitis through TGFβ1. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2015, 35, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Christ, B.; Bruckner, S.; Winkler, S. The therapeutic promise of mesenchymal stem cells for liver restoration. Trends Mol. Med. 2015, 21, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.J.; Gupta, S.; Dhawan, A. Cell therapy for liver disease: From liver transplantation to cell factory. J. Hepatol. 2015, 62, S157–S169. [Google Scholar] [CrossRef] [PubMed]
- Benseler, V.; Obermajer, N.; Johnson, C.L.; Soeder, Y.; Dahlke, M.D.; Popp, F.C. MSC-based therapies in solid organ transplantation. Hepatol. Int. 2014, 8, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Meier, R.P.; Muller, Y.D.; Morel, P.; Gonelle-Gispert, C.; Buhler, L.H. Transplantation of mesenchymal stem cells for the treatment of liver diseases, is there enough evidence? Stem Cell Res. 2013, 11, 1348–1364. [Google Scholar] [CrossRef] [PubMed]
- Stock, P.; Bruckner, S.; Winkler, S.; Dollinger, M.M.; Christ, B. Human bone marrow mesenchymal stem cell-derived hepatocytes improve the mouse liver after acute acetaminophen intoxication by preventing progress of injury. Int. J. Mol. Sci. 2014, 15, 7004–7028. [Google Scholar] [CrossRef] [PubMed]
- Winkler, S.; Borkham-Kamphorst, E.; Stock, P.; Bruckner, S.; Dollinger, M.; Weiskirchen, R.; Christ, B. Human mesenchymal stem cells towards non-alcoholic steatohepatitis in an immunodeficient mouse model. Exp. Cell Res. 2014, 326, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Sokal, E.M.; Stephenne, X.; Ottolenghi, C.; Jazouli, N.; Clapuyt, P.; Lacaille, F.; Najimi, M.; de Lonlay, P.; Smets, F. Liver engraftment and repopulation by in vitro expanded adult derived human liver stem cells in a child with ornithine carbamoyltransferase deficiency. JIMD Rep. 2014, 13, 65–72. [Google Scholar] [PubMed]
- Keating, A. Mesenchymal stromal cells: New directions. Cell Stem Cell 2012, 10, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.H.; Song, F.Q.; Ren, L.N.; Guo, W.Q.; Wang, T.; Feng, Y.X.; Tang, L.J.; Li, K. The multiple functional roles of mesenchymal stem cells in participating in treating liver diseases. J. Cell. Mol. Med. 2015, 19, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Tsolaki, E.; Yannaki, E. Stem cell-based regenerative opportunities for the liver: State of the art and beyond. World J. Gastroenterol. 2015, 21, 12334–12350. [Google Scholar] [CrossRef] [PubMed]
- Squillaro, T.; Peluso, G.; Galderisi, U. Clinical trials with mesenchymal stem cells: An update. Cell Transpl. 2016, 25, 829–848. [Google Scholar] [CrossRef] [PubMed]
- Aurich, H.; Sgodda, M.; Kaltwasser, P.; Vetter, M.; Weise, A.; Liehr, T.; Brulport, M.; Hengstler, J.G.; Dollinger, M.M.; Fleig, W.E.; et al. Hepatocyte differentiation of mesenchymal stem cells from human adipose tissue in vitro promotes hepatic integrationin vivo. Gut 2009, 58, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Aurich, I.; Mueller, L.P.; Aurich, H.; Luetzkendorf, J.; Tisljar, K.; Dollinger, M.M.; Schormann, W.; Walldorf, J.; Hengstler, J.G.; Fleig, W.E.; et al. Functional integration of hepatocytes derived from human mesenchymal stem cells into mouse livers. Gut 2007, 56, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Manuguerra-Gagne, R.; Boulos, P.R.; Ammar, A.; Leblond, F.A.; Krosl, G.; Pichette, V.; Lesk, M.R.; Roy, D.C. Transplantation of mesenchymal stem cells promotes tissue regeneration in a glaucoma model through laser-induced paracrine factor secretion and progenitor cell recruitment. Stem Cells 2013, 31, 1136–1148. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yao, H.L.; Cui, C.B.; Wauthier, E.; Barbier, C.; Costello, M.J.; Moss, N.; Yamauchi, M.; Sricholpech, M.; Gerber, D.; et al. Paracrine signals from mesenchymal cell populations govern the expansion and differentiation of human hepatic stem cells to adult liver fates. Hepatology 2010, 52, 1443–1454. [Google Scholar] [CrossRef] [PubMed]
- Fouraschen, S.G. Support of hepaticr by trophic factors from liver-derived mesenchymal stromal/stem cells. In Animal Models for Stem Cell Therapy; Christ, B., Oerlecke, J., Stock, P., Eds.; Springer: New York, NY, USA, 2014; pp. 89–104. [Google Scholar]
- Parekkadan, B.; van Poll, D.; Megeed, Z.; Kobayashi, N.; Tilles, A.W.; Berthiaume, F.; Yarmush, M.L. Immunomodulation of activated hepatic stellate cells by mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2007, 363, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Li, J.J.; Cao, D.Y.; Li, X.; Zhang, L.Y.; He, Y.; Yue, S.Q.; Wang, D.S.; Dou, K.F. Intravenous injection of mesenchymal stem cells is effective in treating liver fibrosis. World J. Gastroenterol. 2012, 18, 1048–1058. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Kitade, H.; Ni, Y.; Ota, T. Roles of chemokines and chemokine receptors in obesity-associated insulin resistance and nonalcoholic fatty liver disease. Biomolecules 2015, 5, 1563–1579. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, M.J.; Shih, H.; Schafer, R.; Pittenger, M.F. Unraveling the mesenchymal stromal cells’ paracrine immunomodulatory effects. Transfus. Med. Rev. 2016, 30, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Konala, V.B.; Mamidi, M.K.; Bhonde, R.; Das, A.K.; Pochampally, R.; Pal, R. The current landscape of the mesenchymal stromal cell secretome: A new paradigm for cell-free regeneration. Cytotherapy 2016, 18, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Fausto, N.; Campbell, J.S.; Riehle, K.J. Liver regeneration. J. Hepatol. 2012, 57, 692–694. [Google Scholar] [CrossRef] [PubMed]
- Fausto, N.; Campbell, J.S.; Riehle, K.J. Liver regeneration. Hepatology 2006, 43, S45–S53. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, G.K. Liver regeneration after partial hepatectomy: Critical analysis of mechanistic dilemmas. Am. J. Pathol. 2010, 176, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Lawler, P.R.; Lawler, J. Molecular basis for the regulation of angiogenesis by thrombospondin-1 and -2. Cold Spring Harb. Perspect. Med. 2012, 2, a006627. [Google Scholar] [CrossRef] [PubMed]
- Monga, S.P. Role and regulation of β-catenin signaling during physiological liver growth. Gene Expr. 2014, 16, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Liu, C.; Zhou, D.; Zhang, L. TGF-β/SMAD pathway and its regulation in hepatic fibrosis. J. Histochem. Cytochem. 2016, 64, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Schon, H.T.; Weiskirchen, R. Immunomodulatory effects of transforming growth factor-β in the liver. Hepatobiliary Surg. Nutr. 2014, 3, 386–406. [Google Scholar] [PubMed]
- Correia, A.C.; Moonen, J.R.; Brinker, M.G.; Krenning, G. FGF2 inhibits endothelial-mesenchymal transition through microRNA-20a-mediated repression of canonical TGF-β signaling. J. Cell Sci. 2016, 129, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Weiskirchen, R.; Tacke, F. Cellular and molecular functions of hepatic stellate cells in inflammatory responses and liver immunology. Hepatobiliary Surg. Nutr. 2014, 3, 344–363. [Google Scholar] [PubMed]
- Schmidt-Arras, D.; Rose-John, S. IL-6 pathway in the liver: From physiopathology to therapy. J. Hepatol. 2016, 64, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Nijs, J.; Meeus, M.; Versijpt, J.; Moens, M.; Bos, I.; Knaepen, K.; Meeusen, R. Brain-derived neurotrophic factor as a driving force behind neuroplasticity in neuropathic and central sensitization pain: A new therapeutic target? Expert Opin. Ther. Targets 2015, 19, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Fernandez Vallone, V.B.; Romaniuk, M.A.; Choi, H.; Labovsky, V.; Otaegui, J.; Chasseing, N.A. Mesenchymal stem cells and their use in therapy: What has been achieved? Differ. Res. Biol. Divers. 2013, 85, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Phinney, D.G.; Prockop, D.J. Concise review: Mesenchymal stem/multipotent stromal cells: The state of transdifferentiation and modes of tissue repair-current views. Stem Cells 2007, 25, 2896–2902. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.S.; Xin, Z.C.; Dai, J.; Lue, T.F. Commonly used mesenchymal stem cell markers and tracking labels: Limitations and challenges. Histol. Histopathol. 2013, 28, 1109–1116. [Google Scholar] [PubMed]
- Kobolak, J.; Dinnyes, A.; Memic, A.; Khademhosseini, A.; Mobasheri, A. Mesenchymal stem cells: Identification, phenotypic characterization, biological properties and potential for regenerative medicine through biomaterial micro-engineering of their niche. Methods 2016. [Google Scholar] [CrossRef] [PubMed]
- Dan, P.; Velot, E.; Decot, V.; Menu, P. The role of mechanical stimuli in the vascular differentiation of mesenchymal stem cells. J. Cell Sci. 2015, 128, 2415–2422. [Google Scholar] [CrossRef] [PubMed]
- D’souza, N.; Rossignoli, F.; Golinelli, G.; Grisendi, G.; Spano, C.; Candini, O.; Osturu, S.; Catani, F.; Paolucci, P.; Horwitz, E.M.; et al. Mesenchymal stem/stromal cells as a delivery platform in cell and gene therapies. BMC Med. 2015, 13, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christ, B.; Dollinger, M.M. The generation of hepatocytes from mesenchymal stem cells and engraftment into the liver. Curr. Opin. Organ Transpl. 2011, 16, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Kadota, Y.; Yagi, H.; Inomata, K.; Matsubara, K.; Hibi, T.; Abe, Y.; Kitago, M.; Shinoda, M.; Obara, H.; Itano, O.; et al. Mesenchymal stem cells support hepatocyte function in engineered liver grafts. Organogenesis 2014, 10, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Xu, R.; Zhang, Z.; Chen, L.; Shi, M.; Wang, F.S. Implications of the immunoregulatory functions of mesenchymal stem cells in the treatment of human liver diseases. Cell. Mol. Immunol. 2011, 8, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, J.R.; Rosu-Myles, M. Uncovering the secretes of mesenchymal stem cells. Biochimie 2013, 95, 2212–2221. [Google Scholar] [CrossRef] [PubMed]
- Park, C.W.; Kim, K.S.; Bae, S.; Son, H.K.; Myung, P.K.; Hong, H.J.; Kim, H. Cytokine secretion profiling of human mesenchymal stem cells by antibody array. Int. J. Stem Cells 2009, 2, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, I.; Moreno-Caceres, J.; Sanchez, A.; Dooley, S.; Dewidar, B.; Giannelli, G.; Ten Dijke, P.; Consortium, I.-L. TGF-β signaling and liver disease. FEBS J. 2016, 283, 2219–2232. [Google Scholar] [CrossRef] [PubMed]
- Beringer, A.; Noack, M.; Miossec, P. IL-17 in chronic inflammation: From discovery to targeting. Trends Mol. Med. 2016, 22, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, S.; Bonavita, E.; Gentile, S.; Rubino, M.; Laface, I.; Garlanda, C.; Mantovani, A. The long pentraxin PTX3 as a key component of humoral innate immunity and a candidate diagnostic for inflammatory diseases. Int. Arch. Allergy Immunol. 2014, 165, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, S.; Moalli, F.; Ragnarsdottir, B.; Bonavita, E.; Puthia, M.; Riva, F.; Barbati, E.; Nebuloni, M.; Cvetko Krajinovic, L.; Markotic, A.; et al. The humoral pattern recognition molecule PTX3 is a key component of innate immunity against urinary tract infection. Immunity 2014, 40, 621–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusnati, M.; Camozzi, M.; Moroni, E.; Bottazzi, B.; Peri, G.; Indraccolo, S.; Amadori, A.; Mantovani, A.; Presta, M. Selective recognition of fibroblast growth factor-2 by the long pentraxin PTX3 inhibits angiogenesis. Blood 2004, 104, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Fausto, N.; Laird, A.D.; Webber, E.M. Liver regeneration. 2. Role of growth factors and cytokines in hepatic regeneration. FASEB J. 1995, 9, 1527–1536. [Google Scholar] [PubMed]
- Karlmark, K.R.; Wasmuth, H.E.; Trautwein, C.; Tacke, F. Chemokine-directed immune cell infiltration in acute and chronic liver disease. Expert Rev. Gastroenterol. Hepatol. 2008, 2, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Heissig, B.; Dhahri, D.; Eiamboonsert, S.; Salama, Y.; Shimazu, H.; Munakata, S.; Hattori, K. Role of mesenchymal stem cell-derived fibrinolytic factor in tissue regeneration and cancer progression. Cell. Mol. Life Sci. 2015, 72, 4759–4770. [Google Scholar] [CrossRef] [PubMed]
- Blaber, R.; Stylianou, E.; Clayton, A.; Steadman, R. Selective regulation of ICAM-1 and rantes gene expression after ICAM-1 ligation on human renal fibroblasts. J. Am. Soc. Nephrol. 2003, 14, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Kanikarla-Marie, P.; Jain, S.K. Role of hyperketonemia in inducing oxidative stress and cellular damage in cultured hepatocytes and type 1 diabetic rat liver. Cell. Physiol. Biochem. 2015, 37, 2160–2170. [Google Scholar] [CrossRef] [PubMed]
- Jodon de Villeroche, V.; Brouty-Boye, D. Establishment and characterization of atypical fibroblasts from human adult liver contributing to hepatocyte cord-like arrangement. Cell Biol. Int. 2008, 32, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Mars, W.M.; Liu, M.L.; Kitson, R.P.; Goldfarb, R.H.; Gabauer, M.K.; Michalopoulos, G.K. Immediate early detection of urokinase receptor after partial hepatectomy and its implications for initiation of liver regeneration. Hepatology 1995, 21, 1695–1701. [Google Scholar] [PubMed]
- Kozlova, N.; Jensen, J.K.; Chi, T.F.; Samoylenko, A.; Kietzmann, T. PAI-1 modulates cell migration in a LRP1-dependent manner via β-catenin and ERK1/2. Thromb. Haemost. 2015, 113, 988–998. [Google Scholar] [CrossRef] [PubMed]
- Dimova, E.Y.; Kietzmann, T. Metabolic, hormonal and environmental regulation of plasminogen activator inhibitor-1 (PAI-1) expression: Lessons from the liver. Thromb. Haemost. 2008, 100, 992–1006. [Google Scholar] [CrossRef] [PubMed]
- Kruithof, E.K. Regulation of plasminogen activator inhibitor type 1 gene expression by inflammatory mediators and statins. Thromb. Haemost. 2008, 100, 969–975. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Hu, K. Kruppel-like factor 4 inhibits the transforming growth factor-β1-promoted epithelial-to-mesenchymal transition via downregulating plasminogen activator inhibitor-1 in lung epithelial cells. Dis. Markers 2015, 2015, 473742. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.H.; She, H.; Han, Y.P.; Wang, J.; Xiong, S.; Asahina, K.; Tsukamoto, H. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 294, G39–G49. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Sakai, K.; Baba, H.; Sakai, T. Thrombospondin-1 is a novel negative regulator of liver regeneration after partial hepatectomy through transforming growth factor-β1 activation in mice. Hepatology 2012, 55, 1562–1573. [Google Scholar] [CrossRef] [PubMed]
- Starlinger, P.; Haegele, S.; Offensperger, F.; Oehlberger, L.; Pereyra, D.; Kral, J.B.; Schrottmaier, W.C.; Badrnya, S.; Reiberger, T.; Ferlitsch, A.; et al. The profile of platelet α-granule released molecules affects postoperative liver regeneration. Hepatology 2016, 63. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, R.; Nakano, Y.; Ohkohchi, N. Platelet administration via the portal vein promotes liver regeneration in rats after 70% hepatectomy. Ann. Surg. 2011, 253, 759–763. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Park, H.W.; Song, J.H.; Gwak, M.S.; Lee, W.J.; Kim, G.; Lee, S.K.; Ko, J.S. Association between intraoperative platelet transfusion and early graft regeneration in living donor liver transplantation. Ann. Surg. 2015. [Google Scholar] [CrossRef] [PubMed]
- Kir, S.; Beddow, S.A.; Samuel, V.T.; Miller, P.; Previs, S.F.; Suino-Powell, K.; Xu, H.E.; Shulman, G.I.; Kliewer, S.A.; Mangelsdorf, D.J. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science 2011, 331, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Larpthaveesarp, A.; Ferriero, D.M.; Gonzalez, F.F. Growth factors for the treatment of ischemic brain injury (growth factor treatment). Brain Sci. 2015, 5, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Cassiman, D.; Denef, C.; Desmet, V.J.; Roskams, T. Human and rat hepatic stellate cells express neurotrophins and neurotrophin receptors. Hepatology 2001, 33, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.G.; Da Silva, C.A.; Dela Cruz, C.S.; Ahangari, F.; Ma, B.; Kang, M.J.; He, C.H.; Takyar, S.; Elias, J.A. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu. Rev. Physiol. 2011, 73, 479–501. [Google Scholar] [CrossRef] [PubMed]
- Polyzos, S.A.; Kountouras, J.; Mantzoros, C.S. Adipokines in nonalcoholic fatty liver disease. Metab. Clin. Exp. 2015. [Google Scholar] [CrossRef]
- Segers, F.M.; Verdam, F.J.; de Jonge, C.; Boonen, B.; Driessen, A.; Shiri-Sverdlov, R.; Bouvy, N.D.; Greve, J.W.; Buurman, W.A.; Rensen, S.S. Complement alternative pathway activation in human nonalcoholic steatohepatitis. PLoS ONE 2014, 9, e110053. [Google Scholar] [CrossRef] [PubMed]
- Simo, R.; Saez-Lopez, C.; Barbosa-Desongles, A.; Hernandez, C.; Selva, D.M. Novel insights in SHBG regulation and clinical implications. Trends Endocrinol. Metab. 2015, 26, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Le, T.N.; Nestler, J.E.; Strauss, J.F., 3rd; Wickham, E.P., 3rd. Sex hormone-binding globulin and type 2 diabetes mellitus. Trends Endocrinol. Metab. 2012, 23, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Hao, N.B.; Li, C.Z.; Lu, M.H.; Tang, B.; Wang, S.M.; Wu, Y.Y.; Liang, G.P.; Yang, S.M. SDF-1/CXCR4 axis promotes MSCs to repair liver injury partially through trans-differentiation and fusion with hepatocytes. Stem Cells Int. 2015, 2015, 960387. [Google Scholar] [CrossRef] [PubMed]
- Bernhagen, J.; Krohn, R.; Lue, H.; Gregory, J.L.; Zernecke, A.; Koenen, R.R.; Dewor, M.; Georgiev, I.; Schober, A.; Leng, L.; et al. MIF is a noncognate ligand of cxc chemokine receptors in inflammatory and atherogenic cell recruitment. Nat. Med. 2007, 13, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Winkler, S.; Hempel, M.; Bruckner, S.; Mallek, F.; Weise, A.; Liehr, T.; Tautenhahn, H.M.; Bartels, M.; Christ, B. Mouse white adipose tissue-derived mesenchymal stem cells gain pericentral and periportal hepatocyte features after differentiation in vitro, which are preservedin vivo after hepatic transplantation. Acta Physiol. 2015, 215, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M.; et al. String 8—A global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37, D412–D416. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Belinky, F.; Nativ, N.; Stelzer, G.; Zimmerman, S.; Iny Stein, T.; Safran, M.; Lancet, D. Pathcards: Multi-source consolidation of human biological pathways. Database J. Biol. Databases Curation 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Piersma, B.; Bank, R.A.; Boersema, M. Signaling in fibrosis: TGF-β, Wnt, and YAP/TAZ converge. Front. Med. 2015, 2, 59. [Google Scholar] [CrossRef] [PubMed]







| hbmMSC, Undifferentiated—High Abundance Analytes | |||
|---|---|---|---|
| Associated KEGG-Pathway | p-Value | Analytes Involved (Entrez Gene ID) | Analytes Not Found (Entrez Gene ID) |
| p53 signalling pathway | 3.5 × 10−3 | IGFBP-3 (3486) Thromposondin-1 (7057) Serpin E1 (5054) | Angiogenin (283) Chitinase 3-like 1 (1116) MIF (4282) Pentraxin-3 (5806) |
| Cytokine-cytokine receptor interaction | 4.7 × 10−2 | MCP-1 (6347) IL-17A (3605) PDGF-AA (5154) | |
| Complement and coagulation cascade | 9.1 × 10−2 | Serpin E1 (5054) Complement factor D (1675) | |
| hsubMSC, Undifferentiated—High Abundance Analytes | |||
| Associated KEGG-Pathway | p-Value | Analytes Involved (Entrez Gene ID) | Analytes Not Found (Entrez Gene ID) |
| Cytokine-cytokine receptor interaction | 2.4 × 10−3 | MCP-1 (6347) ENA-78 (6374) IL-17A (3605) IL-8 (3576) | Dkk-1 (22,943) Pentraxin-3 (5806) |
| Chemokine signalling pathway | 1.8 × 10−2 | MCP-1 (6347) ENA-78 (6374) IL-8 (3576) | |
| Bladder cancer | 4.9 × 10−2 | IL-8 (3576) Thromposondin-1 (7057) | |
| NOD-like receptor signalling pathway | 7.1 × 10−2 | IL-8 (3576) MCP-1 (6347) | |
| p53 signalling pathway | 7.8 × 10−2 | Thromposondin-1 (7057) Serpin E1 (5054) | |
| hbmMSC, Hepatocytic Differentiated—High Abundance Analytes | |||
|---|---|---|---|
| Associated KEGG-Pathway | p-Value | Analytes Involved (Entrez Gene ID) | Analytes Not Found (Entrez Gene ID) |
| Cytokine-cytokine receptor interaction | 2.4 × 10−11 | MCP-1 (6347) MCP-3 (3654) MIP-3α (6364) RANTES (6352) GRO-α (2919) SDF-1 α (6387) ENA-78 (6374) M-CSF (1435) GM-CSF (1437) IL-17A (3605) IL-6 (3569) IL-8 (3576) VEGF (7422) | Angiogenin (283) Angiopoetin-1 (284) EMMPRIN (682) Chitinase 3-like 1 (1116) Cystatin C (1471) Dkk-1 (22,943) Endoglin (2022) FGF-19 (9965) GDF-15 (9518) IGFBP-2 (3485) MIF (4282) Pentraxin-3 (5806) Resistin (56,729) |
| NOD-like receptor signalling pathway | 3.1 × 10−6 | MCP-1 (6347) RANTES (6352) MCP-3 (6354) GRO-α (2919) IL-6 (3569) IL-8 (3576) | |
| Chemokine signalling pathway | 4.2 × 10−6 | MCP-1 (6347) MIP-3α (6364) RANTES (6352) MCP-3 (6354) GRO-α (2919) SDF-1 α (6387) ENA-78 (6374) IL-8 (3576) | |
| Bladder cancer | 1.2 × 10−2 | IL-8 (3576) Thromposondin-1 (7057) VEGF (7422) | |
| p53 signalling pathway | 2.9 × 10−2 | IGFBP-3 (3486) Serpin E1 (5054) Trombospondin 1 (7057) | |
| Epithelial cell signalling in Heliobacter pylori infection | 2.9 × 10−2 | RANTES (6352) GRO-α (2919) IL-8 (3576) | |
| Complement and coagulation cascade | 2.9 × 10−2 | uPAR (5329) Serpin E1 (5054) Complement factor D (1675) | |
| Hematopoietic cell linage | 4.4 × 10−2 | M-CSF (1435) GM-CSF (1437) IL-6 (3569) | |
| Toll-like receptor signalling pathway | 5.9 × 10−2 | RANTES (6352) IL-6 (3569) IL-8 (3576) | |
| hsubMSC, Hepatocytic Differentiated—High Abundance Analytes | |||
| Associated KEGG-Pathway | p-Value | Analytes Involved (Entrez Gene ID) | Analytes Not Found (Entrez Gene ID) |
| Cytokine-cytokine receptor interaction | 4.0 × 10−6 | MCP-1 (6347) GRO-α (2919) SDF-1 α (6387) ENA-78 (6374) IL-17A (3605) IL-8 (3576) Leptin (3952) VEGF (7422) | Angiogenin (283) EMMPRIN (682) Chitinase 3-like 1 (1116) Cystatin C (1471) Dkk-1 (22,943) DPPIV (1803) Endoglin (2022) GDF-15 (9518) IGFBP-2 (3485) MIF (4282) Pentraxin-3 (2806) FGF-19 (9956) |
| Chemokine signalling pathway | 1.8 × 10−3 | MCP-1 (6347) GRO-α (2919) SDF-1 α (6387) ENA-78 (6374) IL-8 (3576) | |
| Bladder cancer | 6.5 × 10−2 | IL-8 (3576) Thromposondin-1 (7057) VEGF (7422) | |
| NOD-like receptor signalling pathway | 1.4 × 10−2 | MCP-1 (6347) GRO-α (2919) IL-8 (3576) | |
| p53 signalling pathway | 1.7 × 10−2 | IGFBP-3 (3486) Thromposondin-1 (7057) Serpin E1 (5054) | |
| Complement and coagulation cascade | 1.7 × 10−2 | uPAR (5329) Serpin E1 (5054) Complement factor D (1675) | |
| hbmMSC, Undifferentiated—High Abundance Analytes | |||
|---|---|---|---|
| Associated KEGG-Pathway | p-Value | Analytes Involved (Entrez Gene ID) | Predicted Genes (Entrez Gene ID) |
| Complement and coagulation cascades | 3.8 × 10−5 | SerpinE1 (5054) Complement factor D (1675) PLAT (5327) PLG (5340) PLAU (5328) | IGF1 (3479) IGF2 (3481) PLAU (5328) IL-17RA (23,765) CCR2 (1231) PLAT (5327) PLG (5340) PRDM1 (639) PDGFRA (5156) CD47 (961) |
| p53 signalling pathway | 9.3 × 10−4 | IGF1 (3479) IGFBP-3 (3486) Serpin E1 (5054) Thrombospondin 1 (7057) | |
| Prostate cancer | 2.0 × 10−3 | IGF1 (3479) IGF2 (3481) PDGF AA (5154) PDGFRA (5156) | |
| Cytokine-cytokine receptor interaction | 6.0 × 10−3 | PDGF AA (5154) PDGFRA (5156) IL-17 (3506) IL-17RA (23,765) MCP-1 (6347) | |
| Glioma | 1.4 × 10−2 | IGF1 (3479) PDGF AA (5154) PDGFRA (5156) | |
| Melanoma | 1.8 × 10−2 | IGF1 (3479) PDGF AA (5154) PDGFRA (5156) | |
| Focal adhesion | 1.9 × 10−2 | IGF1 (3479) PDGF AA (5154) PDGFRA (5156) Thrombospondin 1 (7057) | |
| hsubMSC, Undifferentiated-High Abundance Analytes | |||
| Associated KEGG-Pathway | p-Value | Analytes Involved (Entrez Gene ID) | Predicted Genes (Entrez Gene ID) |
| Cytokine-cytokine receptor interaction | 6.0 × 10−5 | IL-17 (3506) IL-17RA (23,765) MCP-1 (6347) ENA-78 (6374) IL-8 (3576) CXCR1 (3577) CXCR2 (3579) | LRP5 (4041) LRP6 (4040) CXCR1 (3577) CXCR2 (3579) PLAU (5328) PLAT (5327) PLG (5340) CCR2 (1231) IL-17RA (23,765) RELA (5970) |
| Chemokine signalling pathway | 1.4 × 10−4 | MCP-1 (6347) ENA-78 (6374) IL-8 (3576) CXCR1 (3577) CXCR2 (3579) RELA (5970) | |
| Epithelial cell signalling in Helicobacter pylori infection | 9.3 × 10−4 | IL-8 (3576) CXCR1 (3577) CXCR2 (3579) RELA (5970) | |
| Complement and coagulation cascades | 9.7 × 10−4 | PLAU (5328) PLAT (5327) PLG (5340) Serpin E1 (5054) | |
| NOD-like receptor signalling pathway | 1.4 × 10−2 | MCP-1 (6347) RELA (5970) IL-8 (3576) | |
| Wnt signalling pathway | 7.1 × 10−2 | Dkk-1 (22,943) LRP5 (4041) LRP6 (4040) | |
| hbmMSC, Hepatocytic Differentiated—High Abundance Analytes | |||
|---|---|---|---|
| Associated KEGG-Pathway | p-Value | Involved Analytes (Entrez Gene ID) | Predicted Genes (Entrez Gene ID) |
| Cytokine-cytokine receptor interaction | 1.8 × 10−16 | MCP-1 (6347) MIP-3 α (6364) RANTES (6352) MCP-3 (6354) CCR5 (1234) Gro-α (2919) SDF-1 α (6387) ENA-78 (6374) M-CSF (1435) GM-CSF (1437) CSF2RA (1438) FLT1 (2321) IL-17 (3506) IL-6 (3569) IL-6R (3570) IL-6ST (3572) IL-8 (3576) KDR (3791) VEGF A (7422) | KDR (3791) FLT1 (2321) IGF1 (3479) IL-6R (3570) LRP6 (4040) CCR5 (1234) CSF2RA (1438) IL-6ST (3572) NRP1 (8829) IGF2 (3481) |
| Chemokine signalling pathway | 8.4 × 10−6 | MCP-1 (6347) MIP-3 α (6364) RANTES (6352) MCP-3 (6354) CCR5 (1234) Gro-α (2919) SDF-1 α (6387) ENA-78 (6374) IL-8 (3576) | |
| NOD-like receptor signalling pathway | 2.6 × 10−5 | MCP-1 (6347) RANTES (6352) MCP-3 (6354) Gro-α (2919) IL-6 (3569) IL-8 (3576) | |
| Hematopoietic cell lineage | 1.5 × 10−3 | M-CSF (1435) GM-CSF (1437) CSF2RA (1438) IL-6 (3569) IL-6R (3570) | |
| p53 signalling pathway | 7.2 × 10−3 | IGF1 (3479) IFGBP3 (3486) Serpin E1 (5054) Trombospondin 1 (7057) | |
| JAK-STAT signalling pathway | 9.6 × 10−3 | GM-CSF (1437) CSF2RA (1438) IL-6 (3569) IL-6R (3570) IL-6ST (3572) | |
| Bladder cancer | 2.5 × 10−2 | IL-8 (3576) Thrombospondin-1 (7057) VEGF A (7422) | |
| Focal adhesion | 9.7 × 10−2 | KDR (3791) FLT1 (2321) Thrombospondin 1 (7057) VEGF A (7422) | |
| mTOR signalling pathway | 3.7 × 10−2 | IGF1 (3479) IGF2 (3481) VEGF A (7422) | |
| Pathways in cancer | 4.1 × 10−2 | CFS2RA (1438) FGF-19 (9965) IGF1 (3479) IL-6 (3569) IL-8 (3576) VEGF A (7422) | |
| Epithelial cell signalling in Helicobacter pylori infection | 6.0 × 10−2 | RANTES (6352) Gro-α (2919) IL-8 (3576) | |
| Complement and coagulation cascades | 6.2 × 10−2 | uPAR (5329) Complement factor D (1675) Serpin E1 (5054) | |
| hsubMSC, Hepatocytic Differentiated—High Abundance Analytes | |||
| Associated KEGG-Pathway | p-Value | Analytes Involved (Entrez Gene ID) | Predicted Genes (Entrez Gene ID) |
| Cytokine-cytokine receptor interaction | 1.3 × 10−8 | KDR (3791) FLT1 (2321) MCP-1 (6347) Gro-α (2919) SDF-1 α (6387) ENA-78 (6374) IL-8 (3576) IL-17A (3605) CXCR2 (3579) CXCR4 (7852) Leptin (3952) VEGF A (7422) | KDR (3791) FLT1 (2321) IGF-1 (3479) LRP6 (4040) IGF-2 (3481) NRP1 (8829) PLAU (5328) CXCR4 (7852) HIF1A (3091) CXCR2 (3579) |
| Chemokine signalling pathway | 2.3 × 10−4 | MCP-1 (6347) MIP-3 α (6364) Gro-α (2919) SDF-1 α (6387) ENA-78 (6374) IL-8 (3576) CXCR2 (3579) CXCR4 (7852) | |
| mTOR signalling pathway | 2.0 × 10−3 | HIF1A (3091) IGF1 (3479) IGF2 (3481) VEGF A (7422) | |
| p53 signalling pathway | 4.3 × 10−3 | IGF-1 (3479) IGFBP-3 (3486) Serpin E1 (5054) Thrombospondin 1 (7057) | |
| Complement and coagulation cascades | 4.4 × 10−3 | uPAR (5329) PLAU (5328) Complement factor D (1675) Serpin E1 (5054) | |
| Focal adhesion | 1.6 × 10−2 | KDR (3791) FLT1 (2321) IGF-1 (3479) Thrombospondin 1 (7057) VEGF A (7422) | |
| Bladder cancer | 1.8 × 10−2 | IL-8 (3576) Thrombospondin 1 (7057) VEGF A (7422) | |
| NOD-like receptor signalling pathway | 3.7 × 10−2 | MCP-1 (6347) Gro-α (2919) IL-8 (3576) | |
| Epithel cell signalling in Heliobacter pylori infection | 4.3 × 10−2 | Gro-α (2919) IL-8 (3476) CXCR2 (3579) | |
| Endocytosis | 6.0 × 10−2 | CXCR4 (7852) IL-8 (3576) FLT1 (2321) KDR (3791) | |
| Pathways in cancer | 7.4 × 10−2 | IGF-1 (3479) HIF1A (3091) FGFR2 (2263) FGF-7 (2252) FGF-19 (9965) IL-6 (3569) IL-8 (3576) VEGF A (7422) | |
| TGF-β Pathway-Related Analytes | |||
|---|---|---|---|
| Cell Type | Low Abundance | Medium Abundance | High Abundance |
| hbmMSC, hepatocytic differentiated | BAFF BDNF IL-1α IL-1β IL-2 IL-3 IL-10 IL-13 IL15 IL-16 IL-19 IL-24 MIP-1α/MIP-1β MIP-3β PDGF-AA TARC TGFα | Angiopoetin-2 FGF basic FGF-7 IL-4 IL-11 IL-22 | Angiopoetin-1 FGF-19 GDF-15 IL-17 IL-6 IL-8 MCP-1 MCP-3 MIP-3α RANTES SDF-1α |
| hsubMSC, hepatocytic differentiated | Angiopoetin-1 Angiopoetin-2 BAFF FGF basic IL-1α IL-1β IL-10 IL-11 IL-24 MCP-3 PDGF-AA RANTES TARC | BDNF FGF-7 IL-4 IL-6 IL-22 MIP-3α | FGF-19 GDF-15 IL-8 IL-17A MCP-1 SDF-1α |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winkler, S.; Hempel, M.; Brückner, S.; Tautenhahn, H.-M.; Kaufmann, R.; Christ, B. Identification of Pathways in Liver Repair Potentially Targeted by Secretory Proteins from Human Mesenchymal Stem Cells. Int. J. Mol. Sci. 2016, 17, 1099. https://doi.org/10.3390/ijms17071099
Winkler S, Hempel M, Brückner S, Tautenhahn H-M, Kaufmann R, Christ B. Identification of Pathways in Liver Repair Potentially Targeted by Secretory Proteins from Human Mesenchymal Stem Cells. International Journal of Molecular Sciences. 2016; 17(7):1099. https://doi.org/10.3390/ijms17071099
Chicago/Turabian StyleWinkler, Sandra, Madlen Hempel, Sandra Brückner, Hans-Michael Tautenhahn, Roland Kaufmann, and Bruno Christ. 2016. "Identification of Pathways in Liver Repair Potentially Targeted by Secretory Proteins from Human Mesenchymal Stem Cells" International Journal of Molecular Sciences 17, no. 7: 1099. https://doi.org/10.3390/ijms17071099
APA StyleWinkler, S., Hempel, M., Brückner, S., Tautenhahn, H. -M., Kaufmann, R., & Christ, B. (2016). Identification of Pathways in Liver Repair Potentially Targeted by Secretory Proteins from Human Mesenchymal Stem Cells. International Journal of Molecular Sciences, 17(7), 1099. https://doi.org/10.3390/ijms17071099