IQGAP1 in Podosomes/Invadosomes Is Involved in the Progression of Glioblastoma Multiforme Depending on the Tumor Status
Abstract
:1. Introduction
2. Results
3. Discussion
3.1. IQGAP1 and Mitosis
3.2. IQGAP1 in GBM Astrocytes and GSCs
3.3. IQGAP1 in Macrophages
3.4. IQGAP1 in Neurons
3.5. IQGAP1 in Endothelial Cells
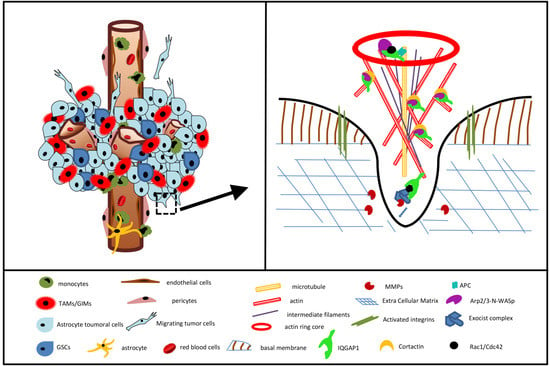
3.6. IQGAP1 in Podosome/Invadopodia
3.7. Final Considerations and Future Research Directions
4. Materials and Methods
4.1. Patients and Tumor Tissue
4.2. Antibodies
4.3. Image Analysis and Statistical Analysis
4.4. Colocalization Analysis
4.5. Double/Triple Immunofluorescence Simultaneous Staining
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ECM | Extracellular matrix |
| EMT | Epithelial-mesenchymal transition |
| FITC | Fluorescein isothiocyanate |
| DAPI | Diamond Anti-fade Mountant |
| GBM | Glioblastoma multiforme |
| GIMs | Glioma-infiltrating myeloid cells |
| Iba1 | Ionized calcium binding adaptor molecule 1 |
| IQGAP1 | IQ motif containing GTPase-activating protein |
| MAP2 | Microtubule-associated protein 2 |
| PCNA | Proliferating cell nuclear antigen |
| TAMs | Tumor associated macrophages |
| TRITC | Tetramethylrhodamine B isothiocyanate |
References
- Lacroix, M.; Abi-Said, D.; Fourney, D.R.; Gokaslan, Z.L.; Shi, W.; DeMonte, F.; Lang, F.F.; McCutcheon, I.E.; Hassenbusch, S.J.; Holland, E.; et al. A multivariate analysis of 416 patients with glioblastoma multiforme: Prognosis, extent of resection, and survival. J. Neurosurg. 2001, 95, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Riemenschneider, M.J.; Reifenberger, G. Molecular neuropathology of gliomas. Int. J. Mol. Sci. 2009, 10, 184–212. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Galanis, E.; Wu, W.; Sarkaria, J.; Chang, S.M.; Colman, H.; Sargent, D.; Reardon, D.A. Incorporation of Biomarker Assessment in Novel Clinical Trial Designs: Personalizing Brain Tumor Treatments. Curr. Oncol. Rep. 2011, 13, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Nicolaides, T.P.; Li, H.; Solomon, D.A.; Hariono, S.; Hashizume, R.; Barkovich, K.; Baker, S.J.; Paugh, B.S.; Jones, C.; Forshew, T.; et al. Targeted therapy for BRAFV600E malignant astrocytoma. Clin. Cancer Res. 2011, 17, 7595–7604. [Google Scholar] [CrossRef] [PubMed]
- Von Daimling, A.; Korshunov, A.; Hartmann, C. The next generation of glioma biomarkers: MGMT methylation, BRAF fusions and IDH1 mutations. Brain Pathol. 2011, 21, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Erickson, J.W.; Cerione, R.A.; Hart, M.J. Identification of an actin cytoskeletal complex that includes IQGAP and the Cdc42 GTPase. J. Biol. Chem. 1997, 272, 24443–24447. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Li, Z.; Sacks, D.B. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol. Cell. Biol. 2005, 25, 7940–7952. [Google Scholar] [CrossRef] [PubMed]
- White, C.D.; Erdemir, H.H.; Sacks, D.B. IQGAP1 and its binding proteins control diverse biological functions. Cell Signal. 2012, 24, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Malarkannan, S.; Awasthi, A.; Rajasekaran, K.; Kumar, P.; Schuldt, K.M.; Bartoszek, A.; Manoharan, N.; Goldner, N.K.; Umhoefer, C.M.; Thakar, M.S. IQGAP1: A regulator of intracellular spacetime relativity. J. Immunol. 2012, 188, 2057–2063. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Sharma, M.; Henderson, B.R. IQGAP1 regulation and roles in cancer. Cell Signal. 2009, 21, 1471–1478. [Google Scholar] [CrossRef] [PubMed]
- White, C.D.; Brown, M.D.; Sacks, D.B. IQGAPs in cancer: A family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009, 583, 1817–1824. [Google Scholar] [CrossRef] [PubMed]
- Nabeshima, K.; Shimao, Y.; Inoue, T.; Koono, M. Immunohistochemical analysis of IQGAP1 expression in human colorectal carcinomas: Its overexpression in carcinomas and association with invasion fronts. Cancer Lett. 2002, 176, 101–109. [Google Scholar] [CrossRef]
- Rotoli, D.; Morales, M.; del Carmen, M.M.; del Pino, G.M.; Gutierrez, R.; Valladares, F.; Avila, J.; Díaz-Flores, L.; Mobasheri, A.; Martin-Vasallo, P. Alterations in IQGAP1 expression and localization in colorectal carcinoma and liver metastases following oxaliplatin-based chemotherapy. Oncol. Lett. 2017, in press. [Google Scholar]
- McDonald, K.L.; O’Sullivan, M.G.; Parkinson, J.F.; Shaw, J.M.; Payne, C.A.; Brewer, J.M.; Young, L.; Reader, D.J.; Wheeler, H.T.; Cook, R.J.; et al. IQGAP1 and IGFBP2: Valuable biomarkers for determining prognosis in glioma patients. J. Neuropathol. Exp. Neurol. 2007, 66, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kim, J.S.; Papadopoulos, J.; Wook, K.S.; Maya, M.; Zhang, F.; He, J.; Fan, D.; Langley, R.; Fidler, I.J. Circulating monocytes expressing CD31: Implications for acute and chronic angiogenesis. Am. J. Pathol. 2009, 174, 1972–1980. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Bao, S. Reciprocal Supportive Interplay between Glioblastoma and Tumor-Associated Macrophages. Cancers (Basel) 2014, 6, 723–740. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Ke, S.Q.; Huang, Z.; Flavahan, W.; Fang, X.; Paul, J.; Wu, L.; Sloan, A.E.; McLendon, R.E.; Li, X.; et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat. Cell Biol. 2015, 17, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ping, Y.F.; Zhang, X.; Bian, X.W. Hostile takeover: Glioma stem cells recruit TAMs to support tumor progression. Cell Stem Cell 2015, 16, 219–220. [Google Scholar] [CrossRef] [PubMed]
- Dell’Albani, P. Stem cell markers in gliomas. Neurochem. Res. 2008, 33, 2407–2415. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Jin, X.; Jung, J.E.; Beck, S.; Kim, H. Cell surface Nestin is a biomarker for glioma stem cells. Biochem. Biophys. Res. Commun. 2013, 433, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, L.; Shen, W.; Liu, Y.; Luo, Y. High expression of VEGF and PI3K in glioma stem cells provides new criteria for the grading of gliomas. Exp. Ther. Med. 2016, 11, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Wiese, C.; Rolletscheck, A.; Kania, G.; Blyszczuk, P.; Tarasov, K.V.; Tarasova, Y.; Wersto, R.P.; Boheler, K.R.; Wobus, A.M. Nestin expression—A property of multi-lineage progenitor cells? Cell. Mol. Life Sci. 2004, 61, 2510–2522. [Google Scholar] [CrossRef] [PubMed]
- Neradil, J.; Veselska, R. Nestin as a marker of cancer stem cells. Cancer Sci. 2015, 106, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, A.; Wickremsekera, A.; Tan, S.T.; Peng, L.; Davis, P.F.; Itinteang, T. Cancer Stem Cell Hierarchy in Glioblastoma Multiforme. Front. Surg. 2016, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Branch, K.M.; Hoshino, D.; Weaver, A.M. Adhesion rings surround invadopodia and promote maturation. Biol. Open 2012, 1, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, T.A.; Lively, S.; Vincent, C.; Schlichter, L.C. Regulation of podosome formation, microglial migration and invasion by Ca2+-signaling molecules expressed in podosomes. J. Neuroinflamm. 2012, 9, 250. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Flores, L.; Gutiérrez, R.; Madrid, J.F.; Varela, H.; Valladares, F.; Acosta, E.; Martín-Vasallo, P.; Díaz-Flores, L., Jr. Pericytes. Morphofunction, interactions and pathology in a quiescent and activated mesenchymal cell niche. Histol. Histopatol. 2009, 4, 909–969. [Google Scholar]
- Wesseling, P.; Schlingemann, R.O.; Rietveld, F.J.; Link, M.; Burger, P.C.; Ruiter, D.J. Early and extensive contribution of pericytes/vascular smooth muscle cells to microvascular proliferation in glioblastoma multiforme: An immuno-light and immuno-electron microscopic study. J. Neuropathol. Exp. Neurol. 1995, 54, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, G.L.; Pfander, B.; Jentsch, S. PCNA, the maestro of the replication fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Sharma, M.; Brocardo, M.G.; Henderson, B.R. IQGAP1 translocates to the nucleus in early S-phase and contributes to cell cycle progression after DNA replication arrest. Int. J. Biochem. Cell Biol. 2011, 43, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Balenci, L.; Clarke, I.D.; Dirks, P.B.; Assard, N.; Ducray, F.; Jouvet, A.; Belin, M.F.; Honnorat, J.; Baudier, J. IQGAP1 protein specifies amplifying cancer cells in glioblastoma multiforme. Cancer Res. 2006, 66, 9074–9082. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.M.; Strasser, A. Is Tumor Growth Sustained by Rare Cancer Stem Cell Or Dominant Clones? Cancer Res. 2008, 68, 4018–4021. [Google Scholar] [CrossRef] [PubMed]
- Coniglio, S.J.; Segall, J.E. Review: Molecular mechanism of microglia stimulated glioblastoma invasion. Matrix Biol. 2013, 32, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Alves, T.R.; Lima, F.R.; Kahn, S.A.; Lobo, D.; Dubois, L.G.; Soletti, R.; Borges, H.; Neto, V.M. Glioblastoma cells: A heterogeneous and fatal tumor interacting with the parenchyma. Life Sci. 2011, 89, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Graeber, M.B. The molecular profile of microglia under the influence of glioma. Neuro Oncol. 2012, 14, 958–978. [Google Scholar] [CrossRef] [PubMed]
- Charles, N.A.; Holland, E.C.; Gilbertson, R.; Glass, R.; Kettenmann, H. The brain tumor microenvironment. Glia 2012, 60, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Rezaie, P.; Patel, K.; Male, D.K. Microglia in the human fetal spinal cord—Patterns of distribution, morphology and phenotype. Dev. Brain Res. 1999, 115, 71–81. [Google Scholar] [CrossRef]
- Ye, X.Z.; Xu, S.L.; Xin, Y.H.; Yu, S.C.; Ping, Y.F.; Chen, L.; Xiao, H.L.; Wang, B.; Yi, L.; Wang, Q.L.; et al. Tumor-associated microglia/macrophages enhance the invasion of glioma stem-like cells via TGF-beta1 signaling pathway. J. Immunol. 2012, 189, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Iwadate, Y. Epithelial-mesenchymal transition in glioblastoma progression. Oncol. Lett. 2016, 11, 1615–1620. [Google Scholar] [CrossRef] [PubMed]
- Rahmathulla, G.; Toms, S.A.; Weil, R.J. The molecular biology of brain metastasis. J. Oncol. 2012, 2012, 723541. [Google Scholar] [CrossRef] [PubMed]
- Huysentruyt, L.C.; Mukherjee, P.; Banerjee, D.; Shelton, L.M.; Seyfried, T.N. Metastatic cancer cells with macrophage properties: Evidence from a new murine tumor model. Int. J. Cancer 2008, 123, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wu, S.; Liang, C.; Lin, Y.; Zou, Y.; Li, K.; Lu, B.; Shu, M.; Huang, Y.; Zhu, W.; et al. Transcriptional upregulation of microtubule-associated protein 2 is involved in the protein kinase A-induced decrease in the invasiveness of glioma cells. Neuro Oncol. 2015, 17, 1578–1588. [Google Scholar] [CrossRef] [PubMed]
- Seano, G.; Chiaverina, G.; Gagliardi, P.A.; di, B.L.; Puliafito, A.; Bouvard, C.; Sessa, R.; Tarone, G.; Sorokin, L.; Helley, D.; et al. Endothelial podosome rosettes regulate vascular branching in tumour angiogenesis. Nat. Cell Biol. 2014, 16, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Gimona, M.; Buccione, R.; Courtneidge, S.A.; Linder, S. Assembly and biological role of podosomes and invadopodia. Curr. Opin. Cell Biol. 2008, 20, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Genot, E.; Gligorijevic, B. Invadosomes in their natural habitat. Eur. J. Cell Biol. 2014, 93, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, D.; Branch, K.M.; Weaver, A.M. Signaling inputs to invadopodia and podosomes. J. Cell Sci. 2013, 126, 2979–2989. [Google Scholar] [CrossRef] [PubMed]
- Poincloux, R.; Lizarraga, F.; Chavrier, P. Matrix invasion by tumour cells: A focus on MT1-MMP trafficking to invadopodia. J. Cell Sci. 2009, 122, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.H.; Jiang, X.J.; Xiao, G.L.; Liu, D.Y.; Yuan, X.R. miR-124a restoration inhibits glioma cell proliferation and invasion by suppressing IQGAP1 and beta-catenin. Oncol. Rep. 2014, 32, 2104–2110. [Google Scholar] [PubMed]
- Rotoli, D.; Morales, M.; del Carmen, M.M.; del Pino, G.M.; Morales, A.; Avila, J.; Martin-Vasallo, P. Expression and localization of the immunophilin FKBP51 in colorectal carcinomas and primary metastases, and alterations following oxaliplatin-based chemotherapy. Oncol. Lett. 2016, 12, 1315–1322. [Google Scholar] [CrossRef] [PubMed]








| Antibody | Marker of |
|---|---|
| Anti-GFAP | Astrocytes |
| Anti-Iba1 | Microglia/Macrophages |
| Anti-PCNA | Proliferative cells |
| Anti-Nestin | Neural stem cells/Glioblastoma stem cells |
| Anti-CD31 | Endothelial cells/Monocyte derived macrophages |
| Anti-Map2 | Mature neurons |
| Anti-β-tubulin | Microtubules |
| Phalloidin-TRITC | F-actin |
| Primary GBM | Patient 1 | Patient 2 | Patient 3 | Patient 4 |
|---|---|---|---|---|
| Intensity localization | +++/++ m,n,c | +++/++ m | +++/++ m,n | +/− m,c |
| Secondary GBM | Patient 5 | Patient 6 | Patient 7 | |
| Intensity localization | +++ m,n | +++/− m,c | +++/++ m,n,c |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rotoli, D.; Pérez-Rodríguez, N.D.; Morales, M.; Maeso, M.D.C.; Ávila, J.; Mobasheri, A.; Martín-Vasallo, P. IQGAP1 in Podosomes/Invadosomes Is Involved in the Progression of Glioblastoma Multiforme Depending on the Tumor Status. Int. J. Mol. Sci. 2017, 18, 150. https://doi.org/10.3390/ijms18010150
Rotoli D, Pérez-Rodríguez ND, Morales M, Maeso MDC, Ávila J, Mobasheri A, Martín-Vasallo P. IQGAP1 in Podosomes/Invadosomes Is Involved in the Progression of Glioblastoma Multiforme Depending on the Tumor Status. International Journal of Molecular Sciences. 2017; 18(1):150. https://doi.org/10.3390/ijms18010150
Chicago/Turabian StyleRotoli, Deborah, Natalia Dolores Pérez-Rodríguez, Manuel Morales, María Del Carmen Maeso, Julio Ávila, Ali Mobasheri, and Pablo Martín-Vasallo. 2017. "IQGAP1 in Podosomes/Invadosomes Is Involved in the Progression of Glioblastoma Multiforme Depending on the Tumor Status" International Journal of Molecular Sciences 18, no. 1: 150. https://doi.org/10.3390/ijms18010150
APA StyleRotoli, D., Pérez-Rodríguez, N. D., Morales, M., Maeso, M. D. C., Ávila, J., Mobasheri, A., & Martín-Vasallo, P. (2017). IQGAP1 in Podosomes/Invadosomes Is Involved in the Progression of Glioblastoma Multiforme Depending on the Tumor Status. International Journal of Molecular Sciences, 18(1), 150. https://doi.org/10.3390/ijms18010150