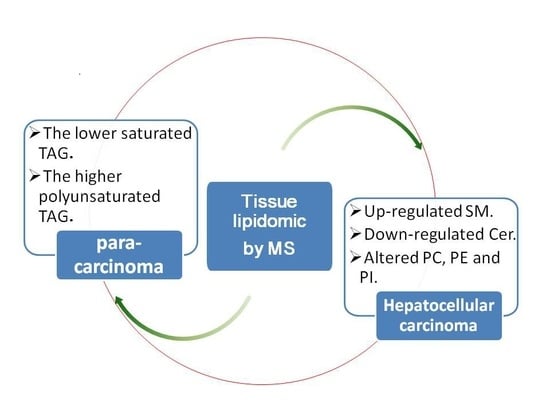
Aberrant Lipid Metabolism in Hepatocellular Carcinoma Revealed by Liver Lipidomics
Abstract
:1. Introduction
2. Results
2.1. Profile Analysis of Lipids in Hepatocellular Carcinoma (HCC) and Para-Carcinoma Tissues by UPLC-ESI-QTOF MS
2.2. Orthogonal Partial Least Squared Discriminant Analysis (OPLS-DA) for Discrimination of HCC (HCC Group) and Para-Carcinoma Tissues (Para-Carcinoma Group)
2.3. Identification of the Aberrant Lipids in HCC Group
2.4. Matrix Assisted Laser Desorption Ionization Mass Spectrometry Imaging (MALDI MSI) Analysis of Human Liver Cancer Tissue
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Tissue Samples
4.3. Lipids Extraction
4.4. Tissue Sectioning
4.5. UPLC-ESI-QTOF Mass Spectrometer
4.6. MALDI-FTICR Mass Spectrometry Imaging
4.7. Data Processing
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| HCC | Hepatocellular carcinoma |
| UPLC-ESI-QTOF MS | Ultra high performance liquid chromatography-electronic spray ionization-QTOF mass spectrometer |
| MALDI-FTICR MS | Matrix assisted laser desorption ionization-fourier transform ion cyclotron resonance mass spectrometer |
| TAG | Triacylglycerol |
| DB | Double bond |
| PC | Phosphatidylcholine |
| PE | Phosphoethanolamine |
| PI | Phosphatidylinositol |
| SM | Sphingomyelin |
| Cer | Ceramide |
| HCV | Hepatitis C virus |
| HBV | Hepatitis B virus |
| NAFLD | Non-alcoholic fatty liver disease |
| NASH | Non-alcoholic steatohepatitis |
| LPC | Lysophosphatidylcholine |
| OPLS-DA | Orthogonal partial least squared discriminant analysis |
| MALDI MSI | Matrix assisted laser desorption ionization mass spectrometry imaging |
| TIC | Total ion chromatogram |
| TNM | Tumor node metastasis |
| C-H TNM | High TNM grade (III, IV) cancer tissue |
| C-L TNM | Low TNM grade (I, II) cancer tissue |
| VIP | Variable importance in projection value |
| RT | Retention time |
| HMDB | Human Metabolome Database |
| FC | Fold change |
| PG | Phosphatidylglycerol |
| AgNPs | Silver nanoparticles |
| H&E | Hematoxylin-eosin staining |
| ESI-MS | Electrospray ionization mass spectrometry |
| UPLC | Ultra high performance liquid chromatography |
| PS | Phosphatidylserine |
| CRC | Colorectal cancer |
| SFA | Saturated fatty acid |
| MUFA | Monounsaturated fatty acid |
| PUFA | Polyunsaturated fatty acid |
| DAG | Diacylglycerol |
| MeOH | Methanol |
| CH3CN | Acetonitrile |
| IPA | Isopropanol |
| PA | Phosphatidic acid |
| PVP | Polyvinyl pyrrolidone |
| AFP | Alpha fetal protein |
| ITO | Indium tin oxide |
| MSE | Multiplexed data acquisition |
| NaTFA | Sodium trifluoroacetate |
| CID | Collision induced dissociation |
References
- Siegel, R.; Ma, J.M.; Zou, Z.H.; Jemal, A. Cancer Statistics, 2014. CA Cancer J. Clin. 2014, 64, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Arzumanyan, A.; Reis, H.; Feitelson, M.A. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat. Rev. Cancer 2013, 13, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Cauchy, F.; Fuks, D.; Belghiti, J. HCC: Current surgical treatment concepts. Langenbeck Arch. Surg. 2012, 397, 681–695. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, T.; Fickert, P.; Magnes, C.; Guelly, C.; Thueringer, A.; Frank, S.; Kratky, D.; Sattler, W.; Reicher, H.; Sinner, F.; et al. Alterations in Lipid Metabolism Mediate Inflammation, Fibrosis, and Proliferation in a Mouse Model of Chronic Cholestatic Liver Injury. Gastroenterology 2012, 142, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Puri, P.; Baillie, R.A.; Wiest, M.M.; Mirshahi, F.; Choudhury, J.; Cheung, O.; Sargeant, C.; Contos, M.J.; Sanyal, A.J. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 2007, 46, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Puri, P.; Wiest, M.M.; Cheung, O.; Mirshahi, F.; Sargeant, C.; Min, H.K.; Contos, M.J.; Sterling, R.K.; Fuchs, M.; Zhou, H.P.; et al. The Plasma Lipidomic Signature of Nonalcoholic Steatohepatitis. Hepatology 2009, 50, 1827–1838. [Google Scholar] [CrossRef] [PubMed]
- Abdelmalek, M.F.; Day, C. Sugar sweetened beverages and fatty liver disease: Rising concern and call to action. J. Hepatol. 2015, 63, 306–308. [Google Scholar] [CrossRef] [PubMed]
- Alwahsh, S.M.; Gebhardt, R. Dietary fructose as a risk factor for non-alcoholic fatty liver disease (NAFLD). Arch. Toxicol. 2017, 91, 1545–1563. [Google Scholar] [CrossRef] [PubMed]
- Park, E.S.; Lee, J.H.; Hong, J.H.; Park, Y.K.; Lee, J.W.; Lee, W.J.; Lee, J.W.; Kim, K.P.; Kim, K.H. Phosphatidylcholine Alteration Identified Using MALDI Imaging MS in HBV-Infected Mouse Livers and Virus-Mediated Regeneration Defects. PLoS ONE 2014, 9, e103955. [Google Scholar] [CrossRef] [PubMed]
- Kessler, S.M.; Simon, Y.; Gemperlein, K.; Gianmoena, K.; Cadenas, C.; Zimmer, V.; Pokorny, J.; Barghash, A.; Helms, V.; van Rooijen, N.; et al. Fatty Acid Elongation in Non-Alcoholic Steatohepatitis and Hepatocellular Carcinoma. Int. J. Mol. Sci. 2014, 15, 5762–5773. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, T.; Atsumi, A.; Matsumori, R.; Nie, T.; Shinozaki, H.; Suzuki-Kemuriyama, N.; Kuba, M.; Nakagawa, Y.; Ishii, K.; Shimada, M.; et al. Elovl6 Promotes Nonalcoholic Steatohepatitis. Hepatology 2012, 56, 2199–2208. [Google Scholar] [CrossRef] [PubMed]
- Muir, K.; Hazim, A.; He, Y.; Peyressatre, M.; Kim, D.Y.; Song, X.L.; Beretta, L. Proteomic and Lipidomic Signatures of Lipid Metabolism in NASH-Associated Hepatocellular Carcinoma. Cancer Res. 2013, 73, 4722–4731. [Google Scholar] [CrossRef] [PubMed]
- Patterson, A.D.; Maurhofer, O.; Beyoglu, D.; Lanz, C.; Krausz, K.W.; Pabst, T.; Gonzalez, F.J.; Dufour, J.F.; Idle, J.R. Aberrant Lipid Metabolism in Hepatocellular Carcinoma Revealed by Plasma Metabolomics and Lipid Profiling. Cancer Res. 2011, 71, 6590–6600. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Ding, Y.; Wang, Y.; Wang, Z.; Yin, X.; Yan, G.; Zhang, L.; Yang, P.; Shen, H. Functional lipidomics: Palmitic acid impairs hepatocellular carcinoma development by modulating membrane fluidity and glucose metabolism. Hepatology 2017, 66, 432–448. [Google Scholar] [CrossRef] [PubMed]
- Wenk, M.R. Lipidomics: New Tools and Applications. Cell 2010, 143, 888–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, Y.; Li, L.; Wei, J.; Xiong, S.; Zhao, Z. High resolution mass spectrometry coupled with multivariate data analysis revealing plasma lipidomic alteration in ovarian cancer in Asian women. Talanta 2016, 150, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, Y.; Liu, J.; Han, J.; Guan, M.; Yang, H.; Lin, Y.; Xiong, S.; Zhao, Z. Electrospray deposition device used to precisely control the matrix crystal to improve the performance of MALDI MSI. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, L.L.; Shangguan, D.H.; Wei, Y.B.; Han, J.J.; Xiong, S.X.; Zhao, Z.W. Ultra-high-performance liquid chromatography electrospray ionization tandem mass spectrometry for accurate analysis of glycerophospholipids and sphingolipids in drug resistance tumor cells. J. Chromatogr. A 2015, 1381, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Freter, C. Lipid Metabolism, Apoptosis and Cancer Therapy. Int. J. Mol. Sci. 2015, 16, 924–949. [Google Scholar] [CrossRef] [PubMed]
- Hummel, J.; Segu, S.; Li, Y.; Irgang, S.; Jueppner, J.; Giavalisco, P. Ultra performance liquid chromatography and high resolution mass spectrometry for the analysis of plant lipids. Front. Plant Sci. 2011, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.W.; Xiao, Y.J.; Elson, P.; Tan, H.Y.; Plummer, S.J.; Berk, M.; Aung, P.P.; Lavery, I.C.; Achkar, J.P.; Li, L.; et al. Plasma lysophosphatidylcholine levels: Potential biomarkers for colorectal cancer. J. Clin. Oncol. 2007, 25, 2696–2701. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Guo, B.; Song, J.W.; Deng, X.L.; Cong, Y.S.; Li, P.F.; Zhao, K.; Liu, L.H.; Xiao, G.; Xu, F.; et al. Plasma choline-containing phospholipids: Potential biomarkers for colorectal cancer progression. Metabolomics 2013, 9, 202–212. [Google Scholar] [CrossRef]
- Van Thiel, D.H.; Alwahsh, S.M.; Ramadori, G. Metabolic Disease and Hepatocellular Carcinoma. In Hepatocellular Carcinoma, Diagnosis and Treatment, 3rd ed.; Brian, I.C., Ed.; Springer: Basel, Switzerland, 2016; pp. 287–301. [Google Scholar]
- Aguilera-Mendez, A.; Alvarez-Delgado, C.; Hernandez-Godinez, D.; Fernandez-Mejia, C. Hepatic Diseases Related to Triglyceride Metabolism. Mini Rev. Med. Chem. 2013, 13, 1691–1699. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Stone, N.J.; Ballantyne, C.; Bittner, V.; Criqui, M.H.; Ginsberg, H.N.; Goldberg, A.C.; Howard, W.J.; Jacobson, M.S.; Kris-Etherton, P.M.; et al. Triglycerides and Cardiovascular Disease A Scientific Statement From the American Heart Association. Circulation 2011, 123, 2292–2333. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.J.; Ginsberg, H.N.; Amarenco, P.; Andreotti, F.; Boren, J.; Catapano, A.L.; Descamps, O.S.; Fisher, E.; Kovanen, P.T.; Kuivenhoven, J.A.; et al. Triglyceride-rich lipoproteins and high-density lipoprotein cholesterol in patients at high risk of cardiovascular disease: Evidence and guidance for management. Eur. Heart J. 2011, 32, 1345–1361. [Google Scholar] [CrossRef] [PubMed]
- Di Angelantonio, E.; Sarwar, N.; Perry, P.; Kaptoge, S.; Ray, K.K.; Thompson, A.; Wood, A.M.; Lewington, S.; Sattar, N.; Packard, C.J.; et al. Major Lipids, Apolipoproteins, and Risk of Vascular Disease. J. Am. Med. Assoc. 2009, 302, 1993–2000. [Google Scholar]
- Alberti, K.; Eckel, R.H.; Grundy, S.M.; Zimmet, P.Z.; Cleeman, J.I.; Donato, K.A.; Fruchart, J.C.; James, W.P.T.; Loria, C.M.; Smith, S.C.; et al. Harmonizing the Metabolic Syndrome: A Joint Interim Statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation 2009, 120, 1640–1645. [Google Scholar] [PubMed]
- Calder, P.C. n-3 polyunsaturated fatty acids and inflammation: From molecular biology to the clinic. Lipids 2003, 38, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.A.; Woodman, R.J.; Burke, V.; Puddey, I.B.; Croft, K.D.; Beilin, L.J. Effect of eicosapentaenoic acid and docosahexaenoic acid on oxidative stress and inflammatory markers in treated-hypertensive type 2 diabetic subjects. Free Radic. Biol. Med. 2003, 35, 772–781. [Google Scholar] [CrossRef]
- Chapkin, R.S.; Davidson, L.A.; Ly, L.; Weeks, B.R.; Lupton, J.R.; McMurray, D.N. Immunomodulatory effects of (n-3) fatty acids: Putative link to inflammation and colon cancer. J. Nutr. 2007, 137, 200S–204S. [Google Scholar] [PubMed]
- Yonezawa, Y.; Hada, T.; Uryu, K.; Tsuzuki, T.; Eitsuka, T.; Miyazawa, T.; Murakami-Nakai, C.; Yoshida, H.; Mizushina, Y. Inhibitory effect of conjugated eicosapentaenoic acid on mammalian DNA polymerase and topoisomerase activities and human cancer cell proliferation. Biochem. Pharmacol. 2005, 70, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Vance, D.E.; Vance, J.E. Phospholipid biosynthesis in eukaryotes. In Biochemistry of Lipids, Lipoproteins and Membranes, 5th ed.; Vance, J.E., Ed.; Elsevier: New York, NY, USA, 2008; pp. 213–244. [Google Scholar]
- Hajj, C.; Becker-Flegler, K.A.; Haimovitz-Friedman, A. Novel mechanisms of action of classical chemotherapeutic agents on sphingolipid pathways. Biol. Chem. 2015, 396, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Li, Z.-Y.; Zhong, Z.; Gates, B.; Xia, Y.; Venkateswaran, S. Synthesis and characterization of stable aqueous dispersions of silver nanoparticles through the Tollens process Electronic supplementary information (ESI) available: Photographs of silver mirror, and of stable dispersions of silver nanoparticles from mixing diluted silvering solutions under sonication at various times. J. Mater. Chem. 2002, 12, 522–527. [Google Scholar]
- Zhao, Z.W.; Xu, Y. An extremely simple method for extraction of lysophospholipids and phospholipids from blood samples. J. Lipid Res. 2010, 51, 652–659. [Google Scholar] [CrossRef] [PubMed]





| Aberrant Lipid | RT (min) | Quasimolecular Ion | m/z | Error (ppm) | p Value | FC | |
|---|---|---|---|---|---|---|---|
| Detected | Theoretical | (C/P) | |||||
| 46:1 TAG | 7.94 | [M + NH4]+ | 794.7217 | 794.7232 | −1.91 | 0.2163 | 1.60 |
| 46:0 TAG | 8.19 | [M + NH4]+ | 796.7369 | 796.7389 | −2.47 | 0.0890 | 1.79 |
| 48:2 TAG | 7.98 | [M + NH4]+ | 820.7380 | 820.7389 | −1.10 | 0.1409 | 1.62 |
| 48:1 TAG | 8.21 | [M + NH4]+ | 822.7521 | 822.7545 | −2.93 | 0.0365 | 1.58 |
| 48:0 TAG | 8.45 | [M + NH4]+ | 824.7680 | 824.7702 | −2.62 | 0.0065 | 2.23 |
| 50:1 TAG | 8.47 | [M + NH4]+ | 850.7843 | 850.7858 | −1.78 | 0.0092 | 1.29 |
| 50:0 TAG | 8.72 | [M + NH4]+ | 852.7995 | 852.8015 | −2.30 | 0.0006 | 3.60 |
| 51:1 TAG | 8.60 | [M + NH4]+ | 864.7989 | 864.8015 | −3.01 | 0.0143 | 1.82 |
| 52:5 TAG | 7.80 | [M + NH4]+ | 870.7511 | 870.7545 | −3.92 | 0.0006 | 0.38 |
| 52:4 TAG | 8.03 | [M + NH4]+ | 872.7698 | 872.7702 | −0.42 | 0.0000 | 0.53 |
| 52:3 TAG | 8.26 | [M + NH4]+ | 874.7859 | 874.7858 | 0.10 | 0.0339 | 0.79 |
| 52:2 TAG | 8.49 | [M + NH4]+ | 876.8004 | 876.8015 | −1.21 | 0.0193 | 0.87 |
| 52:1 TAG | 8.73 | [M + NH4]+ | 878.8148 | 878.8171 | −2.63 | 0.0001 | 1.82 |
| 52:0 TAG | 8.95 | [M + NH4]+ | 880.8298 | 880.8328 | −3.36 | 0.0027 | 4.32 |
| 53:1 TAG | 8.83 | [M + NH4]+ | 892.8300 | 892.8328 | −3.14 | 0.0007 | 4.46 |
| 54:7 TAG | 7.58 | [M + NH4]+ | 894.7510 | 894.7545 | −3.93 | 0.0004 | 0.27 |
| 54:6 TAG | 7.83 | [M + NH4]+ | 896.7674 | 896.7702 | −3.12 | 0.0081 | 0.51 |
| 54:5 TAG | 8.06 | [M + NH4]+ | 898.7831 | 898.7858 | −3.02 | 0.0000 | 0.53 |
| 54:4 TAG | 8.29 | [M + NH4]+ | 900.7993 | 900.8015 | −2.40 | 0.0006 | 0.68 |
| 54:2 TAG | 8.73 | [M + NH4]+ | 904.8303 | 904.8328 | −2.72 | 0.0090 | 1.51 |
| 54:1 TAG | 8.96 | [M + NH4]+ | 906.8451 | 906.8484 | −3.65 | 0.0006 | 3.17 |
| 56:5 TAG | 8.39 | [M + NH4]+ | 926.8146 | 926.8171 | −2.70 | 0.0413 | 2.30 |
| 56:4 TAG | 8.57 | [M + NH4]+ | 928.8298 | 928.8328 | −3.23 | 0.0196 | 1.93 |
| 56:2 TAG | 8.95 | [M + NH4]+ | 932.8607 | 932.8641 | −3.64 | 0.0042 | 3.19 |
| 36:4 DAG | 4.99 | [M + NH4]+ | 634.5394 | 634.5405 | −1.73 | 0.0000 | 0.24 |
| 36:3 DAG | 5.42 | [M + NH4]+ | 636.5544 | 636.5561 | −2.75 | 0.0001 | 0.29 |
| 24:1 SM | 5.60 | [M + H]+ | 813.6817 | 813.6844 | −3.32 | 0.0141 | 1.73 |
| C24:1 1-deoxy Cer | 6.72 | [M + H]+ | 632.6325 | 632.6340 | −2.37 | 0.0003 | 0.48 |
| 22:0 Cer | 6.20 | [M + HCOO]− | 666.6029 | 666.6042 | −1.94 | 0.0005 | 0.65 |
| 23:0 Cer | 6.44 | [M + HCOO]− | 680.6181 | 680.6198 | −2.57 | 0.0000 | 0.55 |
| 24:0 Cer | 6.65 | [M + HCOO]− | 694.6343 | 694.6355 | −1.72 | 0.0012 | 0.65 |
| 16:0/16:1 PC | 3.97 | [M + H]+ | 732.5524 | 732.5538 | −1.89 | 0.0008 | 2.76 |
| 3.89 | [M + HCOO]− | 776.5434 | 776.5447 | −1.68 | 0.0040 | 3.17 | |
| 16:0/16:0 PC | 4.46 | [M + H]+ | 734.5680 | 734.5694 | −1.95 | 0.0315 | 1.43 |
| 4.40 | [M + HCOO]− | 778.5589 | 778.5604 | −1.87 | 0.0047 | 1.55 | |
| 16:0/18:2 PC | 4.00 | [M + HCOO]− | 802.5597 | 802.5604 | −0.82 | 0.0015 | 0.68 |
| 16:0/18:1 PC | 4.45 | [M + HCOO]− | 804.5749 | 804.5760 | −1.37 | 0.0002 | 1.41 |
| 16:0/20:4 PC | 3.98 | [M + H]+ | 782.5687 | 782.5694 | −0.93 | 0.0854 | 0.78 |
| 16:0/20:3 PC | 4.23 | [M + H]+ | 784.5836 | 784.5851 | −1.89 | 0.1719 | 1.31 |
| 16:0/22:6 PC | 3.85 | [M + H]+ | 806.5679 | 806.5694 | −1.86 | 0.0228 | 0.58 |
| 3.80 | [M + HCOO]− | 850.5585 | 850.5604 | −2.18 | 0.0003 | 0.54 | |
| 18:2/18:2 PC | 3.73 | [M + H]+ | 782.5675 | 782.5694 | −2.47 | 0.0007 | 0.32 |
| 18:0/18:1 PC | 5.16 | [M + H]+ | 788.6138 | 788.6164 | −3.27 | 0.0009 | 2.68 |
| 5.08 | [M + HCOO]− | 832.6057 | 832.6073 | −1.93 | 0.0001 | 2.63 | |
| 18:0/20:4 PC | 4.59 | [M + H]+ | 810.5988 | 810.6007 | −2.38 | 0.3424 | 0.84 |
| 18:0/20:3 PC | 4.87 | [M + H]+ | 812.6139 | 812.6164 | −3.05 | 0.0105 | 1.83 |
| 4.81 | [M + HCOO]− | 856.6053 | 856.6073 | −2.34 | 0.0038 | 1.68 | |
| 18:0/22:6 PC | 4.44 | [M + H]+ | 834.5988 | 834.6007 | −2.31 | 0.0407 | 0.63 |
| 16:0/18:1 PE | 3.89 | [M − H]− | 716.5218 | 716.5236 | −2.48 | 0.0039 | 3.33 |
| 16:0/22:6 PE | 3.92 | [M + H]+ | 764.5199 | 764.5225 | −3.38 | 0.0006 | 0.45 |
| 3.87 | [M − H]− | 762.5056 | 762.5079 | −3.05 | 0.0000 | 0.51 | |
| 16:0/22:5 PE | 4.21 | [M + H]+ | 766.5391 | 766.5381 | 1.26 | 0.0236 | 2.37 |
| 4.17 | [M − H]− | 764.5224 | 764.5236 | −1.54 | 0.0133 | 1.68 | |
| 16:0/20:4 PE | 4.03 | [M − H]− | 738.5066 | 738.5079 | −1.76 | 0.2755 | 1.11 |
| 16:1/22:2 PE | 4.95 | [M + H]+ | 770.5683 | 770.5694 | −1.47 | 0.0182 | 2.31 |
| 4.85 | [M − H]− | 768.5532 | 768.5549 | −2.18 | 0.0092 | 1.61 | |
| 18:1/22:5 PE | 4.52 | [M + H]+ | 792.5519 | 792.5538 | −2.37 | 0.0131 | 0.58 |
| 18:0/18:2 PE | 4.00 | [M + H]+ | 742.5380 | 742.5392 | −1.62 | 0.0000 | 0.65 |
| 18:0/18:1 PE | 4.45 | [M + H]+ | 744.5533 | 744.5549 | −2.12 | 0.0002 | 1.41 |
| 18:0/20:3 PE | 5.01 | [M + H]+ | 768.5525 | 768.5549 | −3.10 | 0.0043 | 14.48 |
| 18:0/20:1 PE | 5.08 | [M + H]+ | 772.5841 | 772.5862 | −2.69 | 0.0002 | 2.60 |
| 18:1/22:5 PE | 4.46 | [M + H]+ | 790.5376 | 790.5392 | −2.06 | 0.0056 | 0.65 |
| 18:2/18:2 PG | 2.74 | [M − H]− | 769.5004 | 769.5025 | −2.73 | 0.0000 | 0.31 |
| 18:1/18:2 PG | 3.16 | [M − H]− | 771.5164 | 771.5182 | −2.28 | 0.0000 | 0.37 |
| 18:1/18:1 PG | 3.57 | [M − H]− | 773.5320 | 773.5338 | −2.34 | 0.1034 | 0.51 |
| 16:0/18:2 PI | 3.24 | [M − H]− | 833.5166 | 833.5186 | −2.34 | 0.0056 | 0.64 |
| 16:0/18:1 PI | 3.67 | [M − H]− | 835.5328 | 835.5342 | −1.68 | 0.0205 | 3.75 |
| 18:2/18:2 PI | 3.19 | [M − H]− | 857.5170 | 857.5186 | −1.87 | 0.1499 | 1.23 |
| 18:0/18:2 PI | 3.83 | [M − H]− | 861.5486 | 861.5499 | −1.51 | 0.0000 | 0.48 |
| 18:0/20:3 PI | 4.03 | [M − H]− | 887.5642 | 887.5655 | −1.46 | 0.0256 | 1.52 |
| Factor | No. | % | |
|---|---|---|---|
| Sex | Male | 15 | 75 |
| Female | 5 | 25 | |
| TNM stage | I, II | 13 | 65 |
| III, IV | 7 | 35 | |
| AFP level | <20 ng/mL | 9 | 45 |
| >20 ng/mL | 11 | 55 | |
| vessel invasion | Yes | 9 | 45 |
| No | 11 | 55 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Guan, M.; Lin, Y.; Cui, X.; Zhang, Y.; Zhao, Z.; Zhu, J. Aberrant Lipid Metabolism in Hepatocellular Carcinoma Revealed by Liver Lipidomics. Int. J. Mol. Sci. 2017, 18, 2550. https://doi.org/10.3390/ijms18122550
Li Z, Guan M, Lin Y, Cui X, Zhang Y, Zhao Z, Zhu J. Aberrant Lipid Metabolism in Hepatocellular Carcinoma Revealed by Liver Lipidomics. International Journal of Molecular Sciences. 2017; 18(12):2550. https://doi.org/10.3390/ijms18122550
Chicago/Turabian StyleLi, Zhao, Ming Guan, Yu Lin, Xiao Cui, Yangyang Zhang, Zhenwen Zhao, and Jiye Zhu. 2017. "Aberrant Lipid Metabolism in Hepatocellular Carcinoma Revealed by Liver Lipidomics" International Journal of Molecular Sciences 18, no. 12: 2550. https://doi.org/10.3390/ijms18122550
APA StyleLi, Z., Guan, M., Lin, Y., Cui, X., Zhang, Y., Zhao, Z., & Zhu, J. (2017). Aberrant Lipid Metabolism in Hepatocellular Carcinoma Revealed by Liver Lipidomics. International Journal of Molecular Sciences, 18(12), 2550. https://doi.org/10.3390/ijms18122550