Characteristic Variations and Similarities in Biochemical, Molecular, and Functional Properties of Glyoxalases across Prokaryotes and Eukaryotes
Abstract
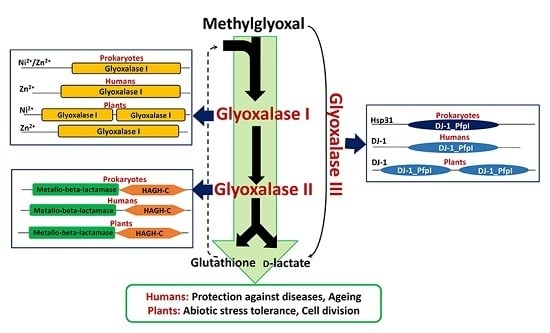
:1. Introduction
2. Metal Ion Dependence Properties of Glyoxalases
3. Presence of Glyoxalase Isoforms in Biological Systems
4. Domain Architecture of Glyoxalases
5. Subcellular Localization Properties of Glyoxalases
6. Kinetics and Regulation of Glyoxalase Enzymatic Activity
7. Structural Variations in Glyoxalase Enzymes
8. Physiological Role of Glyoxalases in Living Systems
9. Functional Diversification in the Plant Glyoxalase Family
10. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Thornalley, P.J.; Waris, S.; Fleming, T.; Santarius, T.; Larkin, S.J.; Winklhofer-Roob, B.M.; Stratton, M.R.; Rabbani, N. Imidazopurinones are markers of physiological genomic damage linked to DNA instability and glyoxalase 1-associated tumour multidrug resistance. Nucleic Acids Res. 2010, 38, 5432–5442. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J. Protein and nucleotide damage by glyoxal and methylglyoxal in physiological systems—Role in ageing and disease. Drug Metabol. Drug Interact. 2008, 23, 125–150. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Sharma, S.; Singla-Pareek, S.L.; Sopory, S.K. Methylglyoxal detoxification in plants: Role of glyoxalase pathway. Ind. J. Plant Physiol. 2016, 21, 377–390. [Google Scholar] [CrossRef]
- Subedi, K.P.; Choi, D.; Kim, I.; Min, B.; Park, C. Hsp31 of Escherichia coli K-12 is glyoxalase III. Mol. Microbiol. 2011, 81, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Kushwaha, H.R.; Hasan, M.R.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Presence of unique glyoxalase III proteins in plants indicates the existence of shorter route for methylglyoxal detoxification. Sci. Rep. 2016, 6, 18358. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Choi, I.G.; Kim, R.; Wang, W.; Jancarik, J.; Yokota, H.; Kim, S.H. Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-Å resolution. Proc. Natl. Acad. Sci. USA 2000, 97, 14079–14084. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Song, J.; Kwon, K.; Jang, S.; Kim, C.; Baek, K.; Kim, J.; Park, C. Human DJ-1 and its homologs are novel glyoxalases. Hum. Mol. Genet. 2012, 21, 3215–3225. [Google Scholar] [CrossRef] [PubMed]
- Sukdeo, N.; Honek, J.K. Pseudomonas aeruginosa contains multiple glyoxalase I-encoding genes from both metal activation classes. Biochim. Biophys. Acta 2007, 1774, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Mustafiz, A.; Ghosh, A.; Tripathi, A.K.; Kaur, C.; Ganguly, A.K.; Bhavesh, N.S.; Tripathi, J.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. A unique Ni2+-dependent and methylglyoxal-inducible rice glyoxalase I possesses a single active site and functions in abiotic stress response. Plant J. 2014, 78, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Batth, R.; Kumari, S.; Mustafiz, A. Arabidopsis thaliana contains both Ni2+ and Zn2+ dependent Glyoxalase I enzymes and ectopic expression of the latter contributes more towards abiotic stress tolerance in E. coli. PLoS ONE 2016, 11, e0159348. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. A glutathione responsive rice glyoxalase II, OsGLYII-2, functions in salinity adaptation by maintaining better photosynthesis efficiency and anti-oxidant pool. Plant J. 2014, 80, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Deswal, R.; Sopory, S.K. Biochemical and immunochemical characterization of Brassica juncea glyoxalase I. Phytochemistry 1998, 49, 2245–2253. [Google Scholar] [CrossRef]
- Clugston, S.L.; Barnard, J.F.; Kinach, R.; Miedema, D.; Ruman, R.; Daub, E.; Honek, J.F. Overproduction and characterization of a dimeric non-zinc glyoxalase I from Escherichia coli: Evidence for optimal activation by nickel ions. Biochemistry 1998, 37, 8754–8763. [Google Scholar] [CrossRef] [PubMed]
- Vickers, T.J.; Greig, N.; Fairlamb, A.H. A trypanothione-dependent glyoxalase I with a prokaryotic ancestry in Leishmania major. Proc. Natl. Acad. Sci. USA 2004, 101, 13186–13191. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Tripathi, A.K.; Nutan, K.K.; Sharma, S.; Ghosh, A.; Tripathi, J.K.; Pareek, A.; Singla-Pareek, S.L.; Sopory, S.K. A nucleus-localized rice glyoxalase I enzyme, OsGLYI-8 functions in the detoxification of methylglyoxal in the nucleus. Plant J. 2016. [Google Scholar] [CrossRef]
- Clugston, S.L.; Yajima, R.; Honek, J.F. Investigation of metal binding and activation of Escherichia coli glyoxalase I: Kinetic, thermodynamic and mutagenesis studies. Biochem. J. 2004, 377, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Ridderstrom, M.; Cameron, A.D.; Jones, T.A.; Mannervik, B. Involvement of an active-site Zn2+ ligand in the catalytic mechanism of human glyoxalase I. J. Biol. Chem. 1998, 273, 21623–21628. [Google Scholar] [CrossRef] [PubMed]
- Schilling, O.; Wenzel, N.; Naylor, M.; Vogel, A.; Crowder, M.; Makaroff, C.; Meyer-Klaucke, W. Flexible metal binding of the metallo-βlactamase domain: Glyoxalase II incorporates iron, manganese, and zinc in vivo. Biochemistry 2003, 42, 11777–11786. [Google Scholar] [CrossRef] [PubMed]
- Limphong, P.; McKinney, R.; Adams, N.; Makaroff, C.; Bennett, B.; Crowder, M. The metal ion requirements of Arabidopsis thaliana Glx2–2 for catalytic activity. J. Biol. Inorg. Chem. 2010, 15, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Marasinghe, G.P.; Sander, I.M.; Bennett, B.; Periyannan, G.; Yang, K.W.; Makaroff, C.A.; Crowder, M.W. Structural studies on a mitochondrial glyoxalase II. J. Biol. Chem. 2005, 280, 40668–40675. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.S.; Barata, L.; Ferreira, A.E.; Romao, S.; Tomas, A.M.; Freire, A.P.; Cordeiro, C. Catalysis and structural properties of Leishmania infantum glyoxalase II: Trypanothione specificity and phylogeny. Biochemistry 2008, 47, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.D.; Ridderstrom, M.; Olin, B.; Mannervik, B. Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue. Structure 1999, 7, 1067–1078. [Google Scholar] [CrossRef]
- Kwon, K.; Choi, D.; Hyun, J.K.; Jung, H.S.; Baek, K.; Park, C. Novel glyoxalases from Arabidopsis thaliana. FEBS J. 2013, 280, 3328–3339. [Google Scholar] [CrossRef]
- Xu, X.M.; Lin, H.; Maple, J.; Bjorkblom, B.; Alves, G.; Larsen, J.P.; Moller, S.G. The Arabidopsis DJ–1a protein confers stress protection through cytosolic SOD activation. J. Cell Sci. 2010, 123, 1644–1651. [Google Scholar] [CrossRef] [PubMed]
- Girotto, S.; Cendron, L.; Bisaglia, M.; Tessari, I.; Mammi, S.; Zanotti, G.; Bubacco, L. DJ-1 is a copper chaperone acting on SOD1 activation. J. Biol. Chem. 2014, 289, 10887–10899. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Vishnoi, A.; Ariyadasa, T.U.; Bhattacharya, A.; Singla-Pareek, S.L.; Sopory, S.K. Episodes of horizontal gene-transfer and gene-fusion led to co-existence of different metal-ion specific glyoxalase I. Sci. Rep. 2013, 3, 3076. [Google Scholar] [CrossRef] [PubMed]
- Aronsson, A.C.; Marmstål, E.; Mannervik, B. Glyoxalase I, a zinc metalloenzyme of mammals and yeast. Biochem. Biophys. Res. Commun. 1978, 81, 1235–1240. [Google Scholar] [CrossRef]
- O’Young, J.; Sukdeo, N.; Honek, J. Escherichia coli glyoxalase II is a binuclear zinc-dependent metalloenzyme. Arch. Biochem. Biophys. 2007, 459, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Mustafiz, A.; Singh, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genom. 2011, 11, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Islam, T. Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 2016, 16, 87. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Lee, J.; Lee, J.Y.; Park, C. Characterization of the Escherichia coli YajL, YhbO and ElbB glyoxalases. FEMS Microbiol. Lett. 2016, 363, fnv239. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Mustafiz, A.; Sarkar, A.K.; Ariyadasa, T.U.; Singla-Pareek, S.L.; Sopory, S.K. Expression of abiotic stress inducible ETHE1-like protein from rice is higher in roots and is regulated by calcium. Physiol. Plant. 2014, 152, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Singla-Pareek, S.L.; Kumar, M.; Pareek, A.; Saxena, M.; Sarin, N.B.; Sopory, S.K. Characterization and functional validation of glyoxalase II from rice. Protein Expr. Purif. 2007, 51, 126–132. [Google Scholar] [CrossRef] [PubMed]
- McCoy, J.G.; Bingman, C.A.; Bitto, E.; Holdorf, M.M.; Makaroff, C.A.; Phillips, G.N. Structure of an ETHE1-like protein from Arabidopsis thaliana. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 964–970. [Google Scholar] [CrossRef] [PubMed]
- Limphong, P.; Nimako, G.; Thomas, P.W.; Fast, W.; Makaroff, C.A.; Crowder, M.W. Arabidopsis thaliana mitochondrial glyoxalase 2–1 exhibits β-lactamase activity. Biochemistry 2009, 48, 8491–8493. [Google Scholar] [CrossRef] [PubMed]
- Frickel, E.M.; Jemth, P.; Widersten, M.; Mannervik, B. Yeast glyoxalase I is a monomeric enzyme with two active sites. J. Biol. Chem. 2001, 276, 1845–1849. [Google Scholar] [CrossRef] [PubMed]
- Bito, A.; Haider, M.; Hadler, I.; Breitenbach, M. Identification and phenotypic analysis of two glyoxalase II encoding genes from Saccharomyces cerevisiae, GLO2 and GLO4, and intracellular localization of the corresponding proteins. J. Biol. Chem. 1997, 272, 21509–21519. [Google Scholar] [CrossRef] [PubMed]
- Miller-Fleming, L.; Antas, P.; Pais, T.F.; Smalley, J.L.; Giorgini, F.; Outeiro, T.F. Yeast DJ-1 superfamily members are required for diauxic-shift reprogramming and cell survival in stationary phase. Proc. Natl. Acad. Sci. USA 2014, 111, 7012–7017. [Google Scholar] [CrossRef] [PubMed]
- Ridderström, M.; Saccucci, F.; Hellman, U.; Bergman, T.; Principato, G.; Mannervik, B. Molecular cloning, heterologous expression, and characterization of human glyoxalase II. J. Biol. Chem. 1996, 271, 319–323. [Google Scholar] [PubMed]
- Shendelman, S.; Jonason, A.; Martinat, C.; Leete, T.; Abeliovich, A. DJ-1 is a redox-dependent molecular chaperone that inhibits α-synuclein aggregate formation. PLoS Biol. 2004, 2, e362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.; Chen, C.; Huang, L.; Yan, J.; Huang, Y. Schizosaccharomyces pombe homologs of human DJ-1 are stationary phase-associated proteins that are involved in autophagy and oxidative stress resistance. PLoS ONE 2015, 10, e0143888. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Su, Y.; Wang, Z.; Chen, C.; Wu, T.; Huang, Y. Identification of glutathione (GSH)-independent glyoxalase III from Schizosaccharomyces pombe. BMC Evol. Biol. 2014, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Bergdoll, M.; Eltis, L.D.; Cameron, A.D.; Dumas, P.; Bolin, J.T. All in the family: Structural and evolutionary relationships among three modular proteins with diverse functions and variable assembly. Protein Sci. 1998, 7, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Deponte, M.; Sturm, N.; Mittler, S.; Harner, M.; Mack, H.; Becker, K. Allosteric coupling of two different functional active sites in monomeric Plasmodium falciparum glyoxalase I. J. Biol. Chem. 2007, 28, 28419–84230. [Google Scholar] [CrossRef] [PubMed]
- Suttisansanee, U.; Honek, J.F. Bacterial glyoxalase enzymes. Semin. Cell Dev. Biol. 2011, 22, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Cookson, M.R. Evolutionary and functional relationships within the DJ1 superfamily. BMC Evol. Biol. 2004, 4, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Ringe, D.; Wilson, M.A.; Ondrechen, M.J. Identification of functional subclasses in the DJ-1 superfamily proteins. PLoS Comput. Biol. 2007, 3, e15. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J. Glyoxalase I–structure, function and a critical role in the enzymatic defence against glycation. Biochem. Soc. Trans. 2003, 31, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, S.C.; Madhubala, R. Glyoxalase I gene deletion mutants of Leishmania donovani exhibit reduced methylglyoxal detoxification. PLoS ONE 2009, 4, e6805. [Google Scholar] [CrossRef] [PubMed]
- Greig, N.; Wyllie, S.; Vickers, T.J.; Fairlamb, A.H. Trypanothione-dependent glyoxalase I in Trypanosoma cruzi. Biochem. J. 2006, 400, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Cordell, P.A.; Futers, T.S.; Grant, P.J.; Pease, R.J. The human hydroxyacylglutathione hydrolase (HAGH) gene encodes both cytosolic and mitochondrial forms of glyoxalase II. J. Biol. Chem. 2004, 279, 28653–28661. [Google Scholar] [CrossRef] [PubMed]
- Urscher, M.; Przyborski, J.; Imoto, M.; Deponte, M. Distinct subcellular localization in the cytosol and apicoplast, unexpected dimerization and inhibition of Plasmodium falciparum glyoxalases. Mol. Microbiol. 2010, 76, 92–195. [Google Scholar] [CrossRef] [PubMed]
- Crowder, M.W.; Maiti, M.K.; Banovic, L.; Makaroff, C.A. Glyoxalase II from A. thaliana requires Zn(II) for catalytic activity. FEBS Lett. 1997, 418, 351–354. [Google Scholar] [CrossRef]
- Maurino, V.G.; Welchen, E.; García, L.; Schmitz, J.; Fuchs, P.; Wagner, S.; Wienstroer, J.; Schertl, P.; Braun, H.P.; Schwarzländer, M.; et al. d-Lactate dehydrogenase links methylglyoxal degradation and electron transport through cytochrome c. Plant Physiol. 2016, 172, 901–912. [Google Scholar] [CrossRef] [PubMed]
- Pallotta, M.L.; Valenti, D.; Iacovino, M.; Passarella, S. Two separate pathways for d-lactate oxidation by Saccharomyces cerevisiae mitochondria which differ in energy production and carrier involvement. Biochim. Biophys. Acta 2004, 1608, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Pallotta, M.L. Mitochondrial involvement to methylglyoxal detoxification: d-Lactate/malate antiporter in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 2012, 102, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shimoji, M.; Thomas, B.; Moore, D.J.; Yu, S.W.; Marupudi, N.I.; Torp, R.; Torgner, I.A.; Ottersen, O.P.; Dawson, T.M.; et al. Mitochondrial localization of the Parkinson’s disease related protein DJ-1: Implications for pathogenesis. Hum. Mol. Gen. 2005, 14, 2063–2073. [Google Scholar] [CrossRef] [PubMed]
- Bankapalli, K.; Saladi, S.; Awadia, S.S.; Goswami, A.V.; Samaddar, M.; D’Silva, P. Robust glyoxalase activity of Hsp31, a ThiJ/DJ-1/PfpI family member protein, is critical for oxidative stress resistance in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 26491–26507. [Google Scholar] [CrossRef] [PubMed]
- Shih, M.J.; Edinger, J.W.; Creighton, D.J. Diffusion-dependent kinetic properties of Glyoxalase I and estimates of the steady-state concentrations of glyoxalase-pathway intermediates in glycolyzing erythrocytes. Eur. J. Biochem. 1997, 244, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Saint-Jean, A.P.; Phillips, K.R.; Creighton, D.J.; Stone, M.J. Active monomeric and dimeric forms of Pseudomonas putida glyoxalase I: Evidence for 3D domain swapping. Biochemistry 1998, 37, 10345–10353. [Google Scholar] [CrossRef] [PubMed]
- Sukdeo, N.; Clugston, S.L.; Daub, E.; Honek, J.F. Distinct classes of glyoxalase I: Metal specificity of the Yersinia pestis, Pseudomonas aeruginosa and Neisseria meningitidis enzymes. Biochem. J. 2004, 384, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Turra, G.L.; Agostini, R.B.; Fauguel, C.M.; Presello, D.A.; Andreo, C.S.; González, J.M.; Campos-Bermudez, V.A. Structure of the novel monomeric glyoxalase I from Zea mays. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 2009–2020. [Google Scholar] [CrossRef] [PubMed]
- Ridderström, M.; Cameron, A.D.; Jones, T.A.; Mannervik, B. Mutagenesis of residue 157 in the active site of human glyoxalase I. Biochem. J. 1997, 328, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Campos-Bermudez, V.A.; Leite, N.R.; Krog, R.; Costa-Filho, A.J.; Soncini, F.C.; Oliva, G.; Vila, A.J. Biochemical and structural characterization of Salmonella typhimurium glyoxalase II: New insights into metal ion selectivity. Biochemistry 2007, 46, 11069–11079. [Google Scholar] [CrossRef] [PubMed]
- Irsch, T.; Krauth-Siegel, R.L. Glyoxalase II of African trypanosomes is trypanothione-dependent. J. Biol. Chem. 2004, 279, 22209–22217. [Google Scholar] [CrossRef] [PubMed]
- Hasim, S.; Hussin, N.A.; Alomar, F.; Bidasee, K.R.; Nickerson, K.W.; Wilson, M.A. A glutathione-independent glyoxalase of the DJ-1 superfamily plays an important role in managing metabolically generated methylglyoxal in Candida albicans. J. Biol. Chem. 2014, 289, 1662–1674. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Lu, T.; Lovett, P.S.; Creighton, D.J. Evidence for a (triosephosphate isomerase-like) “catalytic loop” near the active site of glyoxalase I. J. Biol. Chem. 1995, 2, 12957–12960. [Google Scholar] [CrossRef]
- Sellin, S.; Eriksson, L.E.; Mannervik, B. Fluorescence and nuclear relaxation enhancement studies of the binding of glutathione derivatives to manganese-reconstituted glyoxalase I from human erythrocytes. A model for the catalytic mechanism of the enzyme involving a hydrated metal ion. Biochemistry 1982, 21, 4850–4857. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Sukdeo, N.; Honek, J.F. 15N-1H HSQC NMR evidence for distinct specificity of two active sites in Escherichia coli glyoxalase I. Biochemistry 2008, 47, 13232–13241. [Google Scholar] [CrossRef] [PubMed]
- Campos-Bermudez, V.A.; Morán-Barrio, J.; Costa-Filho, A.J.; Vila, A.J. Metal-dependent inhibition of glyoxalase II: A possible mechanism to regulate the enzyme activity. J. Inorg. Biochem. 2010, 104, 726–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, A.D.; Olin, B.; Ridderström, M.; Mannervik, B.; Jones, T.A. Crystal structure of human glyoxalase I—evidence for gene duplication and 3D domain swapping. EMBO J. 1997, 16, 3386–3395. [Google Scholar] [CrossRef] [PubMed]
- He, M.M.; Clugston, S.L.; Honek, J.F.; Matthews, B.W. Determination of the structure of Escherichia coli glyoxalase I suggests a structural basis for differential metal activation. Biochemistry 2000, 39, 8719–8727. [Google Scholar] [CrossRef] [PubMed]
- Suttisansanee, U.; Lau, K.; Lagishetty, S.; Rao, K.N.; Swaminathan, S.; Sauder, J.M.; Burley, S.K.; Honek, J.F. Structural variation in bacterial glyoxalase I enzymes: Investigation of the metalloenzyme glyoxalase I from Clostridium acetobutylicum. J. Biol. Chem. 2011, 286, 38367–38374. [Google Scholar] [CrossRef] [PubMed]
- Suttisansanee, U.; Ran, Y.; Mullings, K.Y.; Sukdeo, N.; Honek, J.F. Modulating glyoxalase I metal selectivity by deletional mutagenesis: Underlying structural factors contributing to nickel activation profiles. Metallomics 2015, 7, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Carfi, A.; Pares, S.; Duee, E.; Galleni, M.; Duez, C.; Frère, J.M.; Dideberg, O. The 3-D structure of a zinc metallo β-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995, 14, 4914–4921. [Google Scholar] [PubMed]
- Choi, D.; Kim, J.; Ha, S.; Kwon, K.; Kim, E.H.; Lee, H.Y.; Ryu, K.S.; Park, C. Stereospecific mechanism of DJ-1 glyoxalases inferred from their hemithioacetal-containing crystal structures. FEBS J. 2014, 281, 5447–5462. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Tong, L. Crystal structure of human DJ-1, a protein associated with early onset Parkinson’s disease. J. Biol. Chem. 2003, 278, 31372–31379. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Kim, S.J.; Kim, I.K.; Ko, J.; Jeong, C.S.; Kim, G.H.; Park, C.; Kang, S.O.; Suh, P.G.; Lee, H.S.; et al. Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain. J. Biol. Chem. 2003, 278, 44552–44559. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, D.; Kaluarachchi, W.D.; Bellamy, H.D.; White, M.A.; Fox, R.O. The crystal structure of Escherichia coli heat shock protein YedU reveals three potential catalytic active sites. Protein Sci. 2003, 12, 2303–2311. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.A.; Amour, C.V.S.; Collins, J.L.; Ringe, D.; Petsko, G.A. The 1.8-Å resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: A member of the DJ-1/ThiJ/PfpI superfamily. Proc. Natl. Acad. Sci. USA 2004, 101, 1531–1536. [Google Scholar] [CrossRef] [PubMed]
- Morcos, M.; Du, X.; Pfisterer, F.; Hutter, H.; Sayed, A.A.; Thornalley, P.; Ahmed, N.; Baynes, J.; Thorpe, S.; Kukudov, G.; Schlotterer, A. Glyoxalase-1 prevents mitochondrial protein modification and enhances lifespan in Caenorhabditis elegans. Aging Cell 2008, 7, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Fleming, T.H.; Theilen, T.M.; Masania, J.; Wunderle, M.; Karimi, J.; Vittas, S.; Bernauer, R.; Bierhaus, A.; Rabbani, N.; Thornalley, P.J.; et al. Aging-dependent reduction in glyoxalase 1 delays wound healing. Gerontology 2013, 59, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, N.; Xue, M.; Thornalley, P.J. Dicarbonyls and glyoxalase in disease mechanisms and clinical therapeutics. Glycoconj. J. 2016, 33, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Wortmann, M.; Hakimi, M.; Fleming, T.; Peters, A.; Nawroth, P.; Bockler, D.; Dihlmann, S. The role of glyoxalase-1 (Glo-1) in mouse metabolism and atherosclerosis. In Proceedings of the Glyoxalase Centennial: 100 Years of Glyoxalase Research and Emergence of Dicarbonyl Stress, Coventry, UK, 27–29 November 2013.
- Maessen, D.; Brouwers, O.; Miyata, T.; Stehouwer, C.D.A.; Schalkwijk, C.G. Glyoxalase-1 overexpression reduces body weight and adipokine expression, and improves insulin sensitivity in high-fat diet-induced obese mice. In Proceedings of the Diabetologia, Annual Dutch Diabetes Research Meeting, Oosterbeek, The Netherlands, 27–28 November 2014; Springer: New York, NY, USA, 2014; Volume 713. [Google Scholar]
- Zender, L.; Xue, W.; Zuber, J.; Semighini, C.P.; Krasnitz, A.; Ma, B.; Zender, P.; Kubicka, S.; Luk, J.M.; Schirmacher, P.; McCombie, W.R. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 2008, 135, 852–864. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, H.; Mashima, T.; Kizaki, A.; Dan, S.; Hashimoto, Y.; Naito, M.; Tsuruo, T. Glyoxalase I is involved in resistance of human leukemia cells to antitumor agent-induced apoptosis. Blood 2000, 95, 3214–3218. [Google Scholar] [PubMed]
- Williams, R.; Lim, J.E.; Harr, B.; Wing, C.; Walters, R.; Distler, M.G.; Teschke, M.; Wu, C.; Wiltshire, T.; Su, A.I.; et al. A common and unstable copy number variant is associated with differences in Glo1 expression and anxiety-like behavior. PLoS ONE 2009, 4, e4649. [Google Scholar] [CrossRef] [PubMed]
- Distler, M.G.; Plant, L.D.; Sokoloff, G.; Hawk, A.J.; Aneas, I.; Wuenschell, G.E.; Termini, J.; Meredith, S.C.; Nobrega, M.A.; Palmer, A.A. Glyoxalase 1 increases anxiety by reducing GABA A receptor agonist methylglyoxal. J. Clin. Investig. 2012, 122, 2306–2315. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.K.; Singla-Pareek, S.L.; Ray, M.; Reddy, M.K.; Sopory, S.K. Methylglyoxal levels in plants under salinity stress are dependent on glyoxalase I and glutathione. Biochem. Biophys. Res. Commun. 2005, 337, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; Brown, R.L.; Damann, K.E.; Cleveland, T.E. Identification of a maize kernel stress-related protein and its effect on aflatoxin accumulation. Phytopathology 2004, 94, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Singla-Pareek, S.L.; Reddy, M.K.; Sopory, S.K. Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. USA 2003, 100, 14672–14677. [Google Scholar] [CrossRef] [PubMed]
- Singla-Pareek, S.L.; Yadav, S.K.; Pareek, A.; Reddy, M.K.; Sopory, S.K. Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Physiol. 2006, 140, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Xu, J.; Shi, J.; Li, H.; Li, B. Molecular cloning and characterization of a novel glyoxalase I gene TaGLY I in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2010, 37, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, S.; Jamshed, M.; Samuel, M.A. Degradation of glyoxalase I in Brassica napus stigma leads to self-incompatibility response. Nat. Plants 2015, 1, 15185. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Lv, X.; Gao, G.; Li, F.; Li, J.; Qiao, J.; Xu, K.; Chen, B.; Wang, L.; Xiao, X.; et al. Identification and characterization of a glyoxalase I gene in a Rapeseed cultivar with seed thermotolerance. Front. Plant Sci. 2016, 7, 150. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, O.; Guha-Mukherjee, S.; Sopory, S.K. Presence of glyoxalase I in pea. Biochem. Int. 1983, 7, 307–318. [Google Scholar]
- Ramaswamy, O.; Guha-Mukherjee, S.; Sopory, S.K. Correlation of glyoxalase I activity with cell proliferation in Datura callus culture. Plant Cell Rep. 1984, 3, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Bagga, S.; Das, R.; Sopory, S.K. Inhibition of cell proliferation and glyoxalase-I activity by calmodulin inhibitors and lithium in Brassica oleracea. J. Plant Physiol. 1987, 129, 149–153. [Google Scholar] [CrossRef]
- Skoneczna, A.; Miciałkiewicz, A.; Skoneczny, M. Saccharomyces cerevisiae Hsp31p, a stress response protein conferring protection against reactive oxygen species. Free Radic. Biol. Med. 2007, 42, 1409–1420. [Google Scholar] [CrossRef] [PubMed]
- Bonifati, V.; Rizzu, P.; van Baren, M.J.; Schaap, O.; Breedveld, G.J.; Krieger, E.; Dekker, M.C.; Squitieri, F.; Ibanez, P.; Joosse, M.; et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 2003, 299, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Canet-Avilés, R.M.; Wilson, M.A.; Miller, D.W.; Ahmad, R.; McLendon, C.; Bandyopadhyay, S.; Baptista, M.J.; Ringe, D.; Petsko, G.A.; Cookson, M.R. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA 2004, 101, 9103–9108. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Nazarenus, T.J.; Frey, J.L.; Liang, X.; Wilson, M.A.; Stone, J.M. A plant DJ-1 homolog is essential for Arabidopsis thaliana chloroplast development. PLoS ONE 2011, 6, e23731. [Google Scholar] [CrossRef] [PubMed]
- Mulako, I.; Farrant, J.M.; Collett, H.; Illing, N. Expression of Xhdsi-1VOC, a novel member of the vicinal oxygen chelate (VOC) metalloenzyme superfamily, is up-regulated in leaves and roots during desiccation in the resurrection plant Xerophyta humilis (Bak) Dur and Schinz. J. Exp. Bot. 2008, 59, 3885–3901. [Google Scholar] [CrossRef] [PubMed]
- Limphong, P.; Adams, N.E.; Rouhier, M.F.; McKinney, R.M.; Naylor, M.; Bennett, B.; Makaroff, C.A.; Crowder, M.W. Converting GLX2–1 into an active glyoxalase II. Biochemistry 2010, 49, 8228–8236. [Google Scholar] [CrossRef] [PubMed]
- Devanathan, S.; Erban, A.; Perez-Torres Jr, R.; Kopka, J.; Makaroff, C.A. Arabidopsis thaliana glyoxalase 2–1 is required during abiotic stress but is not essential under normal plant growth. PLoS ONE 2014, 9, e95971. [Google Scholar] [CrossRef] [PubMed]
- Holdorf, M.M.; Owen, H.A.; Lieber, S.R.; Yuan, L.; Adams, N.; Dabney-Smith, C.; Makaroff, C.A. Arabidopsis ETHE1 encodes a sulfur dioxygenase that is essential for embryo and endosperm development. Plant Physiol. 2012, 160, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Tiranti, V.; Viscomi, C.; Hildebrandt, T.; di Meo, I.; Mineri, R.; Tiveron, C.; Levitt, M.D.; Prelle, A.; Fagiolari, G.; Rimoldi, M.; et al. Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat. Med. 2009, 15, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Giordano, C.; Viscomi, C.; Orlandi, M.; Papoff, P.; Spalice, A.; Burlina, A.; di Meo, I.; Tiranti, V.; Leuzzi, V.; d’Amati, G.; et al. Morphologic evidence of diffuse vascular damage in human and in the experimental model of ethylmalonic encephalopathy. J. Inherit. Metab. Dis. 2012, 35, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Krüßel, L.; Junemann, J.; Wirtz, M.; Birke, H.; Thornton, J.D.; Browning, L.W.; Poschet, G.; Hell, R.; Balk, J.; Braun, H.P.; et al. The mitochondrial sulfur dioxygenase ethylmalonic encephalopathy protein1 is required for amino acid catabolism during carbohydrate starvation and embryo development in Arabidopsis. Plant Physiol. 2014, 165, 92–104. [Google Scholar] [CrossRef] [PubMed]



| Enzyme Source | GLYI | GLYII | GLYIII | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ni | Zn | Inactive | References | Active | Inactive | References | DJ-1 | Hsp31 | Classification Not Known | References | |
| Escherichia coli | 1 | - | - | [13] | 1 | - | [28] | 1 | 2 | 1 | [4,31] |
| Oryza sativa | 3 | 1 | 7 | [9,26,29] | 2 | 1 | [11,32,33] | 6 | - | - | [5] |
| Arabidopsis thaliana | 2 | 1 | 8 | [10,26,29] | 3 | 2 | [19,20,29,34,35] | 6 | - | - | [23] |
| Glycine max | 8 | 3 | 13 | [30] | 10 | 2 | [30] | N.R. | N.R. | - | |
| Saccharomyces cerevisiae | - | 1 | - | [36] | 2 | - | [37] | 1 | 4 | - | [38] |
| Homo sapiens | - | 1 | - | [17] | 1 | - | [39] | 1 | - | - | [7] |
| Enzyme Source | Protein | Metal | Km (µM) | kcat (s−1) | kcat/Km (M−1·s−1 × 106) | References | ||
|---|---|---|---|---|---|---|---|---|
| Glyoxalase I | ||||||||
| P. aeruginosa | GloA3 | Zn | 287 ± 47 | 787 | 2.8 | [8] | ||
| P. putida | GlxI | Zn | 400 ± 200 | 500 | 1.25 | [60] | ||
| E. coli | GlxI | Ni | 27 ± 0.4 | 338 | 12 | [16] | ||
| Y. pestis | GlxI | Ni | 56 ± 0.6 | 306 | 5.5 | [61] | ||
| P. aeruginosa | GloA1 | Ni | 32 ± 2 | 271 | 8.5 | [8] | ||
| P. aeruginosa | GloA2 | Ni | 21 ± 0 | 247 | 12 | [8] | ||
| N. meningitidis | GlxI | Ni | 45 ± 5 | 204 | 4.5 | [61] | ||
| L. major | GLO1 | Ni | 32 ± 3 | 800 | 25 | [14] | ||
| T. cruzi | TcGLO1 | Ni | 8 ± 0.4 | 161 | 20 | [50] | ||
| P. falciparum | PfGlo1 | Zn | 16 ± 3 * | 103 ± 21 * | 178 * | 285 * | 20.8 ± 2.9 | [44] |
| O. sativa | OsGLYI-11.2 | Ni | 99.8 | 70.96 | 0.71 | [9] | ||
| O. sativa | OsGLYI-8 | Zn | 4.3 ± 1 * | 834 ± 172 * | 22 * | 178 * | 36 ± 8 | [15] |
| A. thaliana | AtGLYI2 | Zn | 786.78 | 137600 | 174.9 | [10] | ||
| A. thaliana | AtGLYI3 | Ni | 45.32 | 728 | 16.08 | [10] | ||
| A. thaliana | AtGLYI6 | Ni | 223.015 | 330 | 1.48 | [10] | ||
| Z. mays | ZmGLX1 | Ni | 56.0 ± 5.0 | N.R. | N.R. | [62] | ||
| S. cerevisiae | GloI | Zn | 410 ± 40 | 1700 | 4.2 | [36] | ||
| H. sapiens | GlxI | Zn | 66 ± 5 | 1500 | 23 | [63] | ||
| Glyoxalase II | ||||||||
| E. coli | GlxII | Zn | 184 ± 22 | 53 | 0.47 | [28] | ||
| S. typhimurium | GloB | Fe-Zn | 241 ± 18 | 394.9 | 1.64 | [64] | ||
| T. brucei | GLX2 | N.R. | ≥3000 | 4.5 | 0.0015 | [65] | ||
| L. infantum | LiGLO2 | Fe-Zn | 324 | 3.52 | 0.0107 | [21] | ||
| O. sativa | OsGLYII-2 | Fe-Zn | 254 ± 12 | 508.33 | 2.0 | [11] | ||
| O. sativa | OsGLYII-3 | N.R. | 61 | 301 | 4.9 | [33] | ||
| A. thaliana | AtGLX2-2 | Fe-Zn | 560 ± 143 | 564 | 1.0 | [19] | ||
| A. thaliana | AtGLX2-5 | Fe-Zn | 391 ± 48 | 129 | 0.33 | [20] | ||
| S. cerevisiae | GLO2 | N.R. | 112 | 979 | 8.7 | [37] | ||
| S. cerevisiae | GLO4 | N.R. | 72.2 | 723 | 10 | [37] | ||
| H. sapiens | GLX2 | Fe-Zn | 187 | 780 | 4.17 | [39] | ||
| Erythrocytes | GLX2 | N.R. | 172 | 755 | 4.39 | [39] | ||
| Bovine liver | GLX2 | N.R. | 190 | 4.37 | 0.023 | [65] | ||
| Glyoxalase III | ||||||||
| Enzyme Source | Protein | Km (mM) | kcat (min−1) | kcat/Km (M−1·min−1 × 105) | References | |||
| E. coli | Hsp31 | 1.43 | 156.9 | 1.1 | [4] | |||
| O. sativa | OsDJ-1C | 0.74 | 2500 | 33.6 | [5] | |||
| A. thaliana | AtDJ-1a | 5.48 | 102 | 0.19 | [23] | |||
| A. thaliana | AtDJ-1b | 4.16 | 154 | 0.37 | [23] | |||
| A. thaliana | AtDJ-1d | 0.1 | 1700 | 170 | [23] | |||
| S. pombe | SpDJ-1 | 10.8 | 85.7 | 0.079 | [42] | |||
| C. albicans | CaGlx3 | 5.5 | 468 | 0.85 | [66] | |||
| S. cerevisiae | Hsp31 | 0.3854 | 150 | 0.578 | [58] | |||
| H. sapiens | HsDJ-1 | 0.6 | 72.38 | 1.21 | [7] | |||
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaur, C.; Sharma, S.; Hasan, M.R.; Pareek, A.; Singla-Pareek, S.L.; Sopory, S.K. Characteristic Variations and Similarities in Biochemical, Molecular, and Functional Properties of Glyoxalases across Prokaryotes and Eukaryotes. Int. J. Mol. Sci. 2017, 18, 250. https://doi.org/10.3390/ijms18040250
Kaur C, Sharma S, Hasan MR, Pareek A, Singla-Pareek SL, Sopory SK. Characteristic Variations and Similarities in Biochemical, Molecular, and Functional Properties of Glyoxalases across Prokaryotes and Eukaryotes. International Journal of Molecular Sciences. 2017; 18(4):250. https://doi.org/10.3390/ijms18040250
Chicago/Turabian StyleKaur, Charanpreet, Shweta Sharma, Mohammad Rokebul Hasan, Ashwani Pareek, Sneh L. Singla-Pareek, and Sudhir K. Sopory. 2017. "Characteristic Variations and Similarities in Biochemical, Molecular, and Functional Properties of Glyoxalases across Prokaryotes and Eukaryotes" International Journal of Molecular Sciences 18, no. 4: 250. https://doi.org/10.3390/ijms18040250
APA StyleKaur, C., Sharma, S., Hasan, M. R., Pareek, A., Singla-Pareek, S. L., & Sopory, S. K. (2017). Characteristic Variations and Similarities in Biochemical, Molecular, and Functional Properties of Glyoxalases across Prokaryotes and Eukaryotes. International Journal of Molecular Sciences, 18(4), 250. https://doi.org/10.3390/ijms18040250