Ectopic Expression of Aeluropus littoralis Plasma Membrane Protein Gene AlTMP1 Confers Abiotic Stress Tolerance in Transgenic Tobacco by Improving Water Status and Cation Homeostasis
Abstract
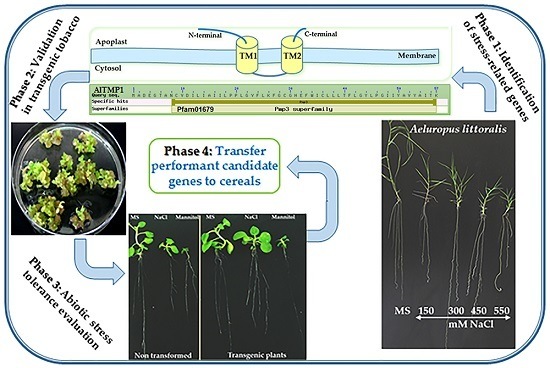
:1. Introduction
2. Results
2.1. Isolation and Sequence Analysis of the AlTMP1 Gene
2.2. AlTMP1 is a Plasma Membrane Protein
2.3. AlTMP1 Transcription Responds to Abiotic Stresses
2.4. AlTMP1 Enhances Abiotic Stress Tolerance in Transgenic Tobacco
2.4.1. Evaluation of Stress Tolerance under In Vitro Conditions
2.4.2. Evaluation of Stress Tolerance under Greenhouse Conditions
AlTMP1 Confers Tolerance to Cold and Heat Stresses
AlTMP1 Confers Tolerance to Continuous Salt and Drought Stresses
2.5. AlTMP1 Regulates the Expression of Some Stress-Related Genes in Transgenic Tobacco
3. Discussion
4. Materials and Methods
4.1. Plant Materials and AlTMP1 Gene Isolation
4.2. Subcellular Localization of GFP::AlTMP1 Fusion Protein
4.3. Q-PCR Analysis of AlTMP1 and Stress-Related Genes
4.4. Generation of Transgenic Tobacco Expressing AlTMP1
4.5. Evaluation of Transgenic Tobacco Tolerance to Abiotic Stresses
4.6. Ion Content Determination
4.7. Membrane Stability Index and Electrolyte Leakage Assay
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Acquaah, G. Principles of Plant Genetics and Breeding; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Gupta, B.A.O.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Zhang, D.F.; Liu, Y.H.; Ying, S.; Shi, Y.S.; Song, Y.C.; Li, Y.; Wang, T.Y. Isolation and characterization of maize PMP3 genes involved in salt stress tolerance. PLoS ONE 2012, 7, e31101. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J.; Munns, R.; Colmer, T.D. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 2015, 115, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Volkov, V. Salinity tolerance in plants. Quantitative approach to ion transport starting from halophytes and stepping to genetic and protein engineering for manipulating ion fluxes. Front. Plant Sci. 2015, 6, 873. [Google Scholar] [CrossRef] [PubMed]
- Bassil, E.; Coku, A.; Blumwald, E. Cellular ion homeostasis: Emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 2012, 63, 5727–5740. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, W.; Rubio, F.; Schroeder, J.I. Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J. 1996, 10, 869–882. [Google Scholar] [CrossRef]
- Uozumi, N.; Kim, E.J.; Rubio, F.; Yamaguchi, T.; Muto, S.; Tsuboi, A.; Bakker, E.P.; Nakamura, T.; Schroeder, J.I. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000, 122, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Demidchik, V.; Tester, M. Sodium fluxes through nonselective cation channels in the plasma membrane of protoplasts from Arabidopsis roots. Plant Physiol. 2002, 128, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.M.; Babourina, O.; Christopher, D.A.; Borsic, T.; Rengel, Z. The cyclic nucleotide-gated channel AtCNGC10 transports Ca2+ and Mg2+ in Arabidopsis. Physiol. Plant. 2010, 139, 303–312. [Google Scholar] [PubMed]
- Navarre, C.; Goffeau, A. Membrane hyperpolarization and salt sensitivity induced by deletion of PMP3, a highly conserved small protein of yeast plasma membrane. EMBO J. 2000, 19, 2515–2524. [Google Scholar] [CrossRef] [PubMed]
- Nylander, M.; Heino, P.; Helenius, E.; Palva, E.T.; Ronne, H.; Welin, B.V. The low-temperature- and salt-induced RCI2A gene of Arabidopsis complements the sodium sensitivity caused by a deletion of the homologous yeast gene SNA1. Plant Mol. Biol. 2001, 45, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Shiozaki, K. The fission yeast stress MAPK cascade regulates the pmp3+ gene that encodes a highly conserved plasma membrane protein. FEBS Lett. 2006, 580, 2409–2413. [Google Scholar] [CrossRef] [PubMed]
- Goddard, N.J.; Dunn, M.A.; Zhang, L.; White, A.J.; Jack, P.L.; Hughes, M.A. Molecular analysis and spatial expression pattern of a low-temperature-specific barley gene, Blt101. Plant Mol. Biol. 1993, 23, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Morsy, M.R.; Almutairi, A.M.; Gibbons, J.; Yun, S.J.; de los Reyes, B.G. The OsLti6 genes encoding low-molecular-weight membrane proteins are differentially expressed in rice cultivars with contrasting sensitivity to low temperature. Gene 2005, 344, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Imai, R.; Koike, M.; Sutoh, K.; Kawakami, A.; Torada, A.; Oono, K. Molecular characterization of a cold-induced plasma membrane protein gene from wheat. Mol. Genet. Genom. 2005, 274, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.H.; Chen, Y.Q.; Wang, Y.; Wang, Z.Z. Molecular cloning and expression of two plasma membrane protein 3 (SmPMP3) genes from Salvia miltiorrhiza. Russ. J. Plant Physiol. 2013, 60, 155–164. [Google Scholar] [CrossRef]
- Medina, J.; Catala, R.; Salinas, J. Developmental and stress regulation of RCI2A and RCI2B, two cold-inducible genes of arabidopsis encoding highly conserved hydrophobic proteins. Plant Physiol. 2001, 125, 1655–1666. [Google Scholar] [CrossRef] [PubMed]
- Inada, M.; Ueda, A.; Shi, W.M.; Takabe, T. A stress-inducible plasma membrane protein 3 (AcPMP3) in a monocotyledonous halophyte, Aneurolepidium chinense, regulates cellular Na+ and K+ accumulation under salt stress. Planta 2005, 220, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Rocha, P.S.C.F. Plant abiotic stress-related RCI2/PMP3s: Multigenes for multiple roles. Planta 2015, 243, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Long, R.C.; Zhang, F.; Li, Z.Y.; Li, M.N.; Cong, L.L.; Kang, J.M.; Zhang, T.J.; Zhao, Z.X.; Sun, Y.; Yang, Q.C. Isolation and functional characterization of salt-stress induced RCI2-like genes from Medicago sativa and Medicago truncatula. J. Plant Res. 2015, 128, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Chang-Qing, Z.; Shunsaku, N.; Shenkui, L.; Tetsuo, T. Characterization of two plasma membrane protein 3 genes (PutPMP3) from the alkali grass, Puccinellia tenuiflora, and functional comparison of the rice homologues, OsLti6a/b from rice. BMB Rep. 2008, 41, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Mitsuya, S.; Taniguchi, M.; Miyake, H.; Takabe, T. Disruption of RCI2A leads to over-accumulation of Na+ and increased salt sensitivity in Arabidopsis thaliana plants. Planta 2005, 222, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Mitsuya, S.; Taniguchi, M.; Miyake, H.; Takabe, T. Overexpression of RCI2A decreases Na+ uptake and mitigates salinity-induced damages in Arabidopsis thaliana plants. Physiol. Plant. 2006, 128, 95–102. [Google Scholar] [CrossRef]
- Liu, B.; Feng, D.R.; Zhang, B.P.; Mu, P.Q.; Zhang, Y.; He, Y.M.; Qi, K.B.; Wang, J.F.; Wang, H.B. Musa paradisica RCI complements AtRCI and confers Na+ tolerance and K+ sensitivity in Arabidopsis. Plant Sci. 2012, 184, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, J.Y.; Kim, S.J.; An, K.S.; An, G.; Kim, S.R. Isolation of cold stress-responsive genes in the reproductive organs, and characterization of the OsLti6b gene from rice (Oryza sativa L.). Plant Cell Rep. 2007, 26, 1097–1110. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Bao, H.; Guo, J.; Jia, W.; Tai, F.; Nie, L.; Jiang, P.; Feng, J.; Lv, S.; Li, Y. Na+/H+ exchanger 1 participates in tobacco disease defence against Phytophthora parasitica var. nicotianae by affecting vacuolar pH and priming the antioxidative system. J. Exp. Bot. 2014, 65, 6107–6122. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Watanabe, T.; Sugano, S.S.; Ueta, R.; Ishihara, R.; Shinozaki, K.; Osakabe, K. Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 2016, 6, 26685. [Google Scholar] [CrossRef] [PubMed]
- Lucas, S.; Dogan, E.; Budak, H. TMPIT1 from wild emmer wheat: First characterisation of a stress-inducible integral membrane protein. Gene 2011, 483, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Zouari, N.; Ben Saad, R.; Legavre, T.; Azaza, J.; Sabau, X.; Jaoua, M.; Masmoudi, K.; Hassairi, A. Identification and sequencing of ESTs from the halophyte grass Aeluropus littoralis. Gene 2007, 404, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Ben Saad, R.; Zouari, N.; Ben Ramdhan, W.; Azaza, J.; Meynard, D.; Guiderdoni, E.; Hassairi, A. Improved drought and salt stress tolerance in transgenic tobacco overexpressing a novel A20/AN1 zinc-finger “AlSAP” gene isolated from the halophyte grass Aeluropus littoralis. Plant Mol. Biol. 2010, 72, 171. [Google Scholar] [CrossRef] [PubMed]
- Ben Saad, R.; Ben Romdhan, W.; Zouari, N.; Azaza, J.; Mieulet, D.; Verdeil, J.-L.; Guiderdoni, E.; Hassairi, A. Promoter of the AlSAP gene from the halophyte grass Aeluropus littoralis directs developmental-regulated, stress-inducible, and organ-specific gene expression in transgenic tobacco. Transg. Res. 2011, 20, 1003–1018. [Google Scholar] [CrossRef] [PubMed]
- Ben Saad, R.; Ben Ramdhan, W.; Zouari, N.; Azaza, J.; Mieulet, D.; Guiderdoni, E.; Ellouz, R.; Hassairi, A. Marker-free transgenic durum wheat cv. Karim expressing the AlSAP gene exhibits a high level of tolerance to salinity and dehydration stresses. Mol. Breed. 2012, 30, 521–533. [Google Scholar] [CrossRef]
- Ben Saad, R.; Fabre, D.; Mieulet, D.; Meynard, D.; Dingkuhn, M.; Al-Doss, A.; Guiderdoni, E.; Hassairi, A. Expression of the Aeluropus littoralis AlSAP gene in rice confers broad tolerance to abiotic stresses through maintenance of photosynthesis. Plant Cell Environ. 2012, 35, 626–643. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Gulick, P.J.; Shen, W.; An, H. ESI3, a stress-induced gene from Lophopyrum elongatum. Plant Physiol. 1994, 104, 799–800. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.R.; Liu, B.; Li, W.Y.; He, Y.M.; Qi, K.B.; Wang, H.B.; Wang, J.F. Over-expression of a cold-induced plasma membrane protein gene (MpRCI) from plantain enhances low temperature-resistance in transgenic tobacco. Environ. Exp. Bot. 2009, 65, 395–402. [Google Scholar] [CrossRef]
- Capel, J.; Jarillo, J.A.; Salinas, J.; Martinez-Zapater, J.M. Two homologous low-temperature-inducible genes from Arabidopsis Encode highly hydrophobic proteins. Plant Physiol. 1997, 115, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Serrano, R.; Rodriguez, P.L. Plants, genes and ions: Workshop on the molecular basis of ionic homeostasis and salt tolerance in plants. EMBO Rep. 2002, 3, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Medina, J.; Ballesteros, M.L.; Salinas, J. Phylogenetic and functional analysis of Arabidopsis RCI2 genes. J. Exp. Bot. 2007, 58, 4333–4346. [Google Scholar] [CrossRef] [PubMed]
- Galvez, A.F.; Gulick, P.J.; Dvorak, J. Characterization of the early stages of genetic salt-stress responses in salt-tolerant Lophopyrum elongatum, salt-sensitive wheat, and their Amphiploid. Plant Physiol. 1993, 103, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Abscisic acid and abiotic stress signaling. Plant Signal Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Giri, B.; Kapoor, R.; Mukerji, K.G. Improved tolerance of Acacia nilotica to salt stress by Arbuscular mycorrhiza, Glomus fasciculatum may be partly related to elevated K/Na ratios in root and shoot tissues. Microb. Ecol. 2007, 54, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guan, C.; Wang, P.; Lv, M.L.; Ma, Q.; Wu, G.Q.; Bao, A.K.; Zhang, J.L.; Wang, S.M. AtHKT1;1 and AtHAK5 mediate low-affinity Na+ uptake in Arabidopsis thaliana under mild salt stress. Plant Growth Regul. 2014, 75, 615–623. [Google Scholar] [CrossRef]
- Bhaskaran, S.; Savithramma, D. Co-expression of Pennisetum glaucum vacuolar Na+/H+ antiporter and Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic tomato. J. Exp. Bot. 2011, 62, 5561–5570. [Google Scholar] [CrossRef] [PubMed]
- Seki, M. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell Online 2001, 13, 61–72. [Google Scholar] [CrossRef]
- Akpinar, B.A.; Avsar, B.; Lucas, S.J.; Budak, H. Plant abiotic stress signaling. Plant Signal Behav. 2012, 7, 1450–1455. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Nebenführ, A. Identifying subcellular protein localization with fluorescent protein fusions after transient expression in onion epidermal cells. Methods Mol. Biol. 2014, 1080, 77–85. [Google Scholar] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Horie, T.; Yoshida, K.; Nakayama, H.; Yamada, K.; Oiki, S.; Shinmyo, A. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 2001, 27, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Kaul, S.; Rounsley, S.; Shea, T.P.; Benito, M.-I.; Town, C.D.; Fujii, C.Y.; Mason, T.; Bowman, C.L.; Barnstead, M.; et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 1999, 402, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.K.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2006, 143, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Tabata, S.; Kaneko, T.; Nakamura, Y.; Kotani, H.; Kato, T.; Asamizu, E.; Miyajima, N.; Sasamoto, S.; Kimura, T.; Hosouchi, T.; et al. Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 2000, 408, 823–826. [Google Scholar] [PubMed]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice,Oryza sativa L. encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Chen, Z.; Du, H.; Liu, Y.; Klessig, D.F. Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J. 1997, 11, 993–1005. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hood, E.E.; Gelvin, S.B.; Melchers, L.S.; Hoekema, A. NewAgrobacterium helper plasmids for gene transfer to plants. Transg. Res. 1993, 2, 208–218. [Google Scholar] [CrossRef]
- Weigel, D.; Glazebrook, J. Transformation of Agrobacterium using electroporation. CSH Protoc. 2006, 2006, 7. [Google Scholar] [CrossRef] [PubMed]
- Horsch, R.B.; Fry, J.; Hoffmann, N.; Neidermeyer, J.; Rogers, S.G.; Fraley, R.T. Leaf disc transformation. In Plant Molecular Biology Manual; Gelvin, S.B., Schilperoort, R.A., Verma, D.P.S., Eds.; Springer: Dordrecht, The Netherlands, 1989; pp. 63–71. [Google Scholar]
- Arnon, D.I. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in β vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hayat, S.; Hasan, S.A.; Yusuf, M.; Hayat, Q.; Ahmad, A. Effect of 28-homobrassinolide on photosynthesis, fluorescence and antioxidant system in the presence or absence of salinity and temperature in Vigna radiata. Environ. Exp. Bot. 2010, 69, 105–112. [Google Scholar] [CrossRef]
- Lutts, S. NaCl-induced senescence in leaves of rice (Oryza sativa L.) cultivars differing in salinity resistance. Ann. Bot. 1996, 78, 389–398. [Google Scholar] [CrossRef]
- Sokal, R.; Rohlf, F. The Principles and Practice of Statistics in Biological Research, 3rd ed.; W. H. Freeman: New York, NY, USA, 1995. [Google Scholar]















© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Romdhane, W.; Ben-Saad, R.; Meynard, D.; Verdeil, J.-L.; Azaza, J.; Zouari, N.; Fki, L.; Guiderdoni, E.; Al-Doss, A.; Hassairi, A. Ectopic Expression of Aeluropus littoralis Plasma Membrane Protein Gene AlTMP1 Confers Abiotic Stress Tolerance in Transgenic Tobacco by Improving Water Status and Cation Homeostasis. Int. J. Mol. Sci. 2017, 18, 692. https://doi.org/10.3390/ijms18040692
Ben Romdhane W, Ben-Saad R, Meynard D, Verdeil J-L, Azaza J, Zouari N, Fki L, Guiderdoni E, Al-Doss A, Hassairi A. Ectopic Expression of Aeluropus littoralis Plasma Membrane Protein Gene AlTMP1 Confers Abiotic Stress Tolerance in Transgenic Tobacco by Improving Water Status and Cation Homeostasis. International Journal of Molecular Sciences. 2017; 18(4):692. https://doi.org/10.3390/ijms18040692
Chicago/Turabian StyleBen Romdhane, Walid, Rania Ben-Saad, Donaldo Meynard, Jean-Luc Verdeil, Jalel Azaza, Nabil Zouari, Lotfi Fki, Emmanuel Guiderdoni, Abdullah Al-Doss, and Afif Hassairi. 2017. "Ectopic Expression of Aeluropus littoralis Plasma Membrane Protein Gene AlTMP1 Confers Abiotic Stress Tolerance in Transgenic Tobacco by Improving Water Status and Cation Homeostasis" International Journal of Molecular Sciences 18, no. 4: 692. https://doi.org/10.3390/ijms18040692
APA StyleBen Romdhane, W., Ben-Saad, R., Meynard, D., Verdeil, J. -L., Azaza, J., Zouari, N., Fki, L., Guiderdoni, E., Al-Doss, A., & Hassairi, A. (2017). Ectopic Expression of Aeluropus littoralis Plasma Membrane Protein Gene AlTMP1 Confers Abiotic Stress Tolerance in Transgenic Tobacco by Improving Water Status and Cation Homeostasis. International Journal of Molecular Sciences, 18(4), 692. https://doi.org/10.3390/ijms18040692