Strategies towards Improved Feed Efficiency in Pigs Comprise Molecular Shifts in Hepatic Lipid and Carbohydrate Metabolism
Abstract
:1. Introduction
2. Results
2.1. Affected Phenotypic Traits Due to Residual Feed Intake (RFI) Classification
2.2. Hepatic Gene Expression Pattern
2.3. Verification of Microarray Results
3. Discussion
3.1. Fatty Acid Concentrations in Feed Efficiency (FE)-Divergent Pigs
3.2. Carbohydrate and Protein Metabolism in FE-Divergent Pigs
3.3. FYN as a Putative Hub Molecule Regulating FE
3.4. Implication on Systemic Integrity of FE-Divergent Pigs
4. Materials and Methods
4.1. Animals, Feed Conversion Testing and Sampling
4.2. Physiological Parameters and Hormones in Serum
4.3. Lipid Extraction and Fatty Acid Profiling
4.4. RNA Isolation
4.5. Microarray Analysis
4.6. Quantitative Real-Time PCR (RT-qPCR)
4.7. Phenotype Data Analyses
4.8. Transcript Data Analyses
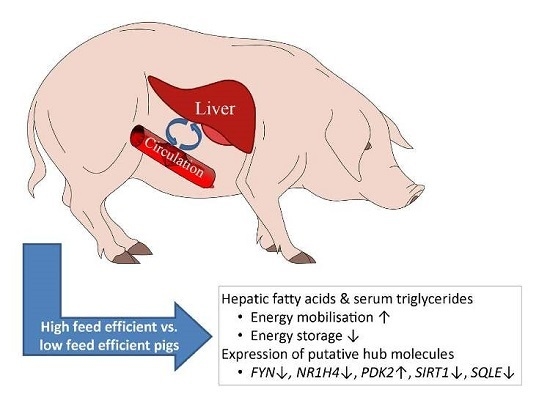
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| FE | Feed efficiency |
| RFI | Residual feed intake |
| FCR | Feed conversion ratio |
| ADG | Average daily weight gain |
| ADFI | Average daily feed intake |
| SFA | Saturated fatty acids |
| MUFA | Monounsaturated fatty acids |
| PUFA | Polyunsaturated fatty acids |
References
- Patience, J.F.; Rossoni-Serão, M.C.; Gutiérrez, N.A. A review of feed efficiency in swine: Biology and application. J. Anim. Sci. Biotechnol. 2015, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.; Szyszka, O.; Stoddart, K.; Edwards, S.; Kyriazakis, I. Animal and management factors influencing grower and finisher pig performance and efficiency in European systems: A meta-analysis. Animal 2015, 9, 1210–1220. [Google Scholar] [CrossRef] [PubMed]
- Fix, J.S.; Cassady, J.P.; van Heugten, E.; Hanson, D.J.; See, M.T. Differences in lean growth performance of pigs sampled from 1980 and 2005 commercial swine fed 1980 and 2005 representative feeding programs. Livest. Sci. 2010, 128, 108–114. [Google Scholar] [CrossRef]
- Do, D.N.; Strathe, A.B.; Jensen, J.; Mark, T.; Kadarmideen, H.N. Genetic parameters for different measures of feed efficiency and related traits in boars of three pig breeds. J. Anim. Sci. 2013, 91, 4069–4079. [Google Scholar] [CrossRef] [PubMed]
- Dodson, M.V.; Hausman, G.J.; Guan, L.; Du, M.; Rasmussen, T.P.; Poulos, S.P.; Mir, P.; Bergen, W.G.; Fernyhough, M.E.; McFarland, D.C. Lipid metabolism, adipocyte depot physiology and utilization of meat animals as experimental models for metabolic research. Int. J. Biol. Sci. 2010, 6, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Jakobsdottir, G.; Xu, J.; Molin, G.; Ahrne, S.; Nyman, M. High-fat diet reduces the formation of butyrate, but increases succinate, inflammation, liver fat and cholesterol in rats, while dietary fibre counteracts these effects. PLoS ONE 2013, 8, e80476. [Google Scholar] [CrossRef] [PubMed]
- Kerr, B.J.; Kellner, T.A.; Shurson, G.C. Characteristics of lipids and their feeding value in swine diets. J. Anim. Sci. Biotechnol. 2015, 6, 30. [Google Scholar] [CrossRef] [PubMed]
- Grubbs, J.; Dekkers, J.; Huff-Lonergan, E.; Tuggle, C.; Lonergan, S. Identification of potential serum biomarkers to predict feed efficiency in young pigs. J. Anim. Sci. 2016, 94, 1482–1492. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, H.; Billon, Y.; Brossard, L.; Faure, J.; Gatellier, P.; Gondret, F.; Labussière, E.; Lebret, B.; Lefaucheur, L.; Le Floch, N. Review: Divergent selection for residual feed intake in the growing pig. Animal 2017, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Le Naou, T.; Le Floch, N.; Louveau, I.; Gilbert, H.; Gondret, F. Metabolic changes and tissue responses to selection on residual feed intake in growing pigs. J. Anim. Sci. 2012, 90, 4771–4780. [Google Scholar] [CrossRef] [PubMed]
- Madeira, M.S.; Pires, V.M.; Alfaia, C.M.; Lopes, P.A.; Martins, S.V.; Pinto, R.M.; Prates, J.A. Restriction of dietary protein does not promote hepatic lipogenesis in lean or fatty pigs. Br. J. Nutr. 2016, 115, 1339–1351. [Google Scholar] [CrossRef] [PubMed]
- Kasprzyk, A.; Tyra, M.; Babicz, M. Fatty acid profile of pork from a local and a commercial breed. AAB 2015, 58, 379. [Google Scholar] [CrossRef]
- Gondret, F.; Louveau, I.; Mourot, J.; Duclos, M.; Lagarrigue, S.; Gilbert, H.; van Milgen, J. Dietary energy sources affect the partition of body lipids and the hierarchy of energy metabolic pathways in growing pigs differing in feed efficiency. J. Anim. Sci. 2014, 92, 4865–4877. [Google Scholar] [CrossRef] [PubMed]
- Vigors, S.; Sweeney, T.; O’Shea, C.; Kelly, A.; O’Doherty, J. Pigs that are divergent in feed efficiency, differ in intestinal enzyme and nutrient transporter gene expression, nutrient digestibility and microbial activity. Animal 2016, 10, 1848–1855. [Google Scholar] [CrossRef] [PubMed]
- Jégou, M.; Gondret, F.; Vincent, A.; Tréfeu, C.; Gilbert, H.; Louveau, I. Whole blood transcriptomics is relevant to identify molecular changes in response to genetic selection for feed efficiency and nutritional status in the pig. PLoS ONE 2016, 11, e0146550. [Google Scholar] [CrossRef] [PubMed]
- Lkhagvadorj, S.; Qu, L.; Cai, W.; Couture, O.P.; Barb, C.R.; Hausman, G.J.; Nettleton, D.; Anderson, L.L.; Dekkers, J.C.; Tuggle, C.K. Gene expression profiling of the short-term adaptive response to acute caloric restriction in liver and adipose tissues of pigs differing in feed efficiency. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 298, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Faure, J.; Lefaucheur, L.; Bonhomme, N.; Ecolan, P.; Meteau, K.; Coustard, S.M.; Kouba, M.; Gilbert, H.; Lebret, B. Consequences of divergent selection for residual feed intake in pigs on muscle energy metabolism and meat quality. Meat Sci. 2013, 93, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.; Patience, J.F.; Lonergan, S.M.; JM Dekkers, C.; Gabler, N.K. Improved nutrient digestibility and retention partially explains feed efficiency gains in pigs selected for low residual feed intake. J. Anim. Sci. 2012, 90, 164–166. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, D.; de Lange, C.; Antonick, T.; Dekkers, J.; Pendleton, A.; Rakhshandeh, A. Effect of divergent selection for residual feed intake on whole body protein turnover in growing gilts fed either adequate or lysine deficient diets. J. Anim. Sci. 2016, 94, 108–109. [Google Scholar] [CrossRef]
- Miar, Y.; Plastow, G.; Bruce, H.; Moore, S.; Manafiazar, G.; Kemp, R.; Charagu, P.; Huisman, A.; van Haandel, B.; Zhang, C. Genetic and phenotypic correlations between performance traits with meat quality and carcass characteristics in commercial crossbred pigs. PLoS ONE 2014, 9, e110105. [Google Scholar] [CrossRef] [PubMed]
- Lefaucher, L.; Lebret, B.; Ecolan, P.; Galian, M.; Damon, M.; Louveau, I.; Prunier, A.; Sellier, P.; Gilbert, H. Divergent selection on “residual feed intake” in pigs: Impact on growth performance, muscle compositional traits and meat quality. AAB 2008, 40, 83–84. [Google Scholar]
- Tizioto, P.C.; Coutinho, L.L.; Decker, J.E.; Schnabel, R.D.; Rosa, K.O.; Oliveira, P.S.; Souza, M.M.; Mourão, G.B.; Tullio, R.R.; Chaves, A.S. Global liver gene expression differences in Nelore steers with divergent residual feed intake phenotypes. BMC Genom. 2015, 16, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grubbs, J.K.; Fritchen, A.; Huff-Lonergan, E.; Dekkers, J.C.; Gabler, N.K.; Lonergan, S.M. Divergent genetic selection for residual feed intake impacts mitochondria reactive oxygen species production in pigs. J. Anim. Sci. 2013, 91, 2133–2140. [Google Scholar] [CrossRef] [PubMed]
- Louveau, I.; Vincent, A.; Tacher, S.; Gilbert, H.; Gondret, F. Increased expressions of genes and proteins involved in mitochondrial oxidation and antioxidant pathway in adipose tissue of pigs selected for a low residual feed intake. J. Anim. Sci. 2016, 94, 5042–5054. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, B.; West, J.A.; Koulman, A. A review of odd-chain fatty acid metabolism and the role of pentadecanoic acid (c15: 0) and heptadecanoic acid (c17: 0) in health and disease. Molecules 2015, 20, 2425–2444. [Google Scholar] [CrossRef] [PubMed]
- Dannenberger, D.; Nuernberg, K.; Nuernberg, G.; Priepke, A. Impact of dietary protein level and source of polyunsaturated fatty acids on lipid metabolism-related protein expression and fatty acid concentrations in porcine tissues. J. Agric. Food Chem. 2014, 62, 12453–12461. [Google Scholar] [CrossRef] [PubMed]
- Ballester, M.; Revilla, M.; Puig-Oliveras, A.; Marchesi, J.; Castello, A.; Corominas, J.; Fernandez, A.; Folch, J. Analysis of the porcine APOA2 gene expression in liver, polymorphism identification and association with fatty acid composition traits. Anim. Genet. 2016, 47, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Xu, S.; Maitland-Toolan, K.A.; Sato, K.; Jiang, B.; Ido, Y.; Lan, F.; Walsh, K.; Wierzbicki, M.; Verbeuren, T.J. Sirt1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J. Biol. Chem. 2008, 283, 20015–20026. [Google Scholar] [CrossRef] [PubMed]
- Purushotham, A.; Schug, T.T.; Xu, Q.; Surapureddi, S.; Guo, X.; Li, X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009, 9, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Lonergan, S.M.; Huff-Lonergan, E.; Rowe, L.; Kuhlers, D.; Jungst, S. Selection for lean growth efficiency in Duroc pigs influences pork quality. J. Anim. Sci. 2001, 79, 2075–2085. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Lu, Y.; Li, X.Y. Farnesoid X receptor: A master regulator of hepatic triglyceride and glucose homeostasis. Acta Pharmacol. Sin. 2015, 36, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Loewenstein, W.R. Junctional intercellular communication and the control of growth. Biochim. Biophys. Acta 1979, 560, 1–65. [Google Scholar] [CrossRef]
- Vatish, M.; Yamada, E.; Pessin, J.E.; Bastie, C.C. Fyn kinase function in lipid utilization: A new upstream regulator of ampk activity? Arch. Physiol. Biochem. 2009, 115, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Bastie, C.C.; Zong, H.; Xu, J.; Busa, B.; Judex, S.; Kurland, I.J.; Pessin, J.E. Integrative metabolic regulation of peripheral tissue fatty acid oxidation by the src kinase family member fyn. Cell Metab. 2007, 5, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Boddicker, N.; Gabler, N.; Spurlock, M.; Nettleton, D.; Dekkers, J. Effects of ad libitum and restricted feeding on early production performance and body composition of Yorkshire pigs selected for reduced residual feed intake. Animal 2011, 5, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Young, J.; Bergsma, R.; Knol, E.; Patience, J.; Dekkers, J. Effect of selection for residual feed intake during the grow/finish phase of production on sow reproductive performance and lactation efficiency. J. Anim. Sci. 2016, 94, 4120–4132. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, T.; Lejeune, A.; Moloney, A.P.; Monahan, F.J.; Mc Gettigan, P.; Downey, G.; Park, S.D.; Ryan, M.T. The application of transcriptomic data in the authentication of beef derived from contrasting production systems. BMC Genom. 2016, 17, 746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Te Pas, M.F.; Koopmans, S.-J.; Kruijt, L.; Calus, M.P.; Smits, M.A. Plasma proteome profiles associated with diet-induced metabolic syndrome and the early onset of metabolic syndrome in a pig model. PLoS ONE 2013, 8, e73087. [Google Scholar] [CrossRef] [PubMed]
- Grundy, S.M.; Brewer, H.B.; Cleeman, J.I.; Smith, S.C.; Lenfant, C. Definition of metabolic syndrome. Circulation 2004, 109, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Kokta, T.; Dodson, M.; Gertler, A.; Hill, R. Intercellular signaling between adipose tissue and muscle tissue. Domest. Anim. Endocrinol. 2004, 27, 303–331. [Google Scholar] [CrossRef] [PubMed]
- Oppenheimer, J.H.; Schwartz, H.L.; Lane, J.T.; Thompson, M.P. Functional relationship of thyroid hormone-induced lipogenesis, lipolysis, and thermogenesis in the rat. J. Clin. Investig. 1991, 87, 125. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Jiang, Y.; Meltzer, P.; Yen, P.M. Thyroid hormone regulation of hepatic genes in vivo detected by complementary DNA microarray. Mol. Endocrinol. 2000, 14, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Seitz, H. Rapid and direct stimulation of hepatic gluconeogenesis by l-triiodothyronine (t3) in the isolated-perfused rat liver. Life Sci. 1980, 27, 827–835. [Google Scholar] [CrossRef]
- McCormack, U.M.; Curião, T.; Buzoianu, S.G.; Prieto, M.L.; Ryan, T.; Varley, P.; Crispie, F.; Magowan, E.; Metzler-Zebeli, B.U.; Berry, D. Exploring a possible link between the intestinal microbiota and feed efficiency in pigs. Appl. Environ. Microbiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Dannenberger, D.; Nuernberg, K.; Nuernberg, G.; Priepke, A. Different dietary protein and pufa interventions alter the fatty acid concentrations, but not the meat quality, of porcine muscle. Nutrients 2012, 4, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Angulo, J.; Hiller, B.; Olivera, M.; Mahecha, L.; Dannenberger, D.; Nuernberg, G.; Losand, B.; Nuernberg, K. Dietary fatty acid intervention of lactating cows simultaneously affects lipid profiles of meat and milk. J. Sci. Food Agric. 2012, 92, 2968–2974. [Google Scholar] [CrossRef] [PubMed]
- Freeman, T.C.; Ivens, A.; Baillie, J.K.; Beraldi, D.; Barnett, M.W.; Dorward, D.; Downing, A.; Fairbairn, L.; Kapetanovic, R.; Raza, S. A gene expression atlas of the domestic pig. BMC Biol. 2012, 10, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kauffmann, A.; Gentleman, R.; Huber, W. Arrayqualitymetrics—A bioconductor package for quality assessment of microarray data. Bioinformatics 2009, 25, 415–416. [Google Scholar] [CrossRef] [PubMed]
- Bourgon, R.; Gentleman, R.; Huber, W. Independent filtering increases detection power for high-throughput experiments. Proc. Natl. Acad. Sci. USA 2010, 107, 9546–9551. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
| Item | Unit | High-FE (Mean ± SE) | Low-FE (Mean ± SE) | p-Value |
|---|---|---|---|---|
| Performance (n = 24) | ||||
| Body weight | kg | 94.96 ± 2.14 | 91.13 ± 3.56 | 0.311 |
| ADG (day 70–day 140) | kg/d | 0.98 ± 0.02 | 0.94 ± 0.04 | 0.653 |
| ADFI (day 70–day 140) | kg/d | 1.90 ± 0.04 | 2.25 ± 0.06 | <0.001 |
| FCR (day 70–day 140) | kg/kg | 1.94 ± 0.03 | 2.40 ± 0.06 | <0.001 |
| RFI | kg | −0.20 ± 0.02 | 0.21 ± 0.03 | <0.001 |
| Backfat | mm | 4.78 ± 0.23 | 4.78 ± 0.31 | 0.650 |
| Liver weight | kg | 1.82 ± 0.08 | 1.65 ± 0.08 | 0.028 |
| Blood parameters (n = 24) | ||||
| Albumin | g/dL | 4.52 ± 0.11 | 4.24 ± 0.11 | 0.181 |
| Glucose | mg/dL | 145.67 ± 29.26 | 110.25 ± 12.05 | 0.183 |
| GGT | U/L | 49.83 ± 5.74 | 47.25 ± 3.99 | 0.600 |
| GPT | U/L | 41.50 ± 1.89 | 37.58 ± 2.07 | 0.175 |
| GOT | U/L | 49.08 ± 4.59 | 41.50 ± 3.39 | 0.175 |
| Urea | mg/dL | 10.89 ± 0.97 | 10.37 ± 0.44 | 0.155 |
| LDH | U/L | 179.42 ± 15.41 | 165.00 ± 15.19 | 0.408 |
| Triglyceride | mg/dL | 39.92 ± 5.58 | 24.75 ± 3.66 | 0.001 |
| Total cholesterol | mg/dL | 107.92 ± 5.45 | 107.83 ± 6.81 | 0.739 |
| Total protein | g/dL | 6.44 ± 0.11 | 6.56 ± 0.18 | 0.916 |
| Amylase | U/L | 828.17 ± 55.49 | 805.92 ± 55.36 | 0.553 |
| Lipase | U/L | 51.75 ± 7.05 | 50.08 ± 6.76 | 0.859 |
| Hormones (n = 12) | ||||
| Triiodothyronine (T3) | ng/mL | 0.59 ± 0.09 | 0.47 ± 0.06 | 0.269 |
| Thyroxine (T4) | ng/mL | 15.62 ± 1.60 | 15.67 ± 2.70 | 0.262 |
| Item | High-FE (Mean ± SE) | Low-FE (Mean ± SE) | p-Value |
|---|---|---|---|
| Sum Fatty Acid Concentrations | |||
| Fat content (%) | 2.36 ± 0.08 | 2.57 ± 0.09 | <0.001 |
| SFA 1 | 912.04 ± 31.63 | 994.21 ± 34.12 | <0.001 |
| MUFA 2 | 285.67 ± 10.68 | 315.63 ± 23.76 | 0.111 |
| PUFA 3 | 1157.42 ± 49.23 | 1261.92 ± 44.00 | <0.001 |
| n-3 PUFA 4 | 151.04 ± 6.27 | 167.11 ± 6.77 | 0.007 |
| n-6 PUFA 5 | 1006.38 ± 43.81 | 1094.81 ± 40.86 | 0.001 |
| Fatty Acid Concentrations | |||
| C10:0 | 2.05 ± 0.18 | 2.22 ± 0.23 | 0.375 |
| C12:0 | 1.46 ± 0.08 | 1.58 ± 0.08 | 0.265 |
| C13:0 | 0.40 ± 0.0 | 0.36 ± 0.02 | 0.028 |
| Fatty Acid Concentrations | |||
| C14:0 | 6.24 ± 0.29 | 7.08 ± 0.98 | 0.333 |
| C15:0 | 4.26 ± 0.29 | 3.70 ± 0.26 | 0.029 |
| C16:0 | 297.99 ± 10.10 | 328.30 ± 17.03 | 0.008 |
| C17:0 | 30.49 ± 2.44 | 27.89 ± 2.57 | 0.627 |
| C18:0 | 546.46 ± 27.46 | 597.87 ± 22.54 | 0.002 |
| C16:1 cis-9 | 10.64 ± 0.82 | 12.32 ± 1.76 | 0.225 |
| C18:1 cis-9 | 223.17 ± 8.75 | 247.07 ± 20.32 | 0.130 |
| C18:1 cis-11 | 31.95 ± 1.60 | 33.16 ± 1.97 | 0.222 |
| C18:1 trans-9 | 4.07 ± 0.22 | 4.20 ± 0.24 | 0.342 |
| C18:1 trans-11 | 1.85 ± 0.05 | 1.79 ± 0.10 | 0.259 |
| C20:1 cis-11 | 4.01 ± 0.18 | 4.22 ± 0.18 | 0.094 |
| C18:2 n-6 | 397.90 ± 23.34 | 460.20 ± 26.03 | <0.001 |
| C18:3 n-3 | 10.25 ± 0.93 | 14.13 ± 2.29 | 0.059 |
| C18:3 n-6 | 4.31 ± 0.19 | 5.24 ± 0.49 | 0.003 |
| C20:2 n-6 | 12.55 ± 0.92 | 14.23 ± 0.78 | 0.011 |
| C20:3 n-6 | 26.11 ± 2.19 | 29.58 ± 1.90 | 0.081 |
| C20:4 n-6 | 535.06 ± 20.46 | 551.75 ± 32.88 | 0.264 |
| C20:5 n-3 | 21.78 ± 1.64 | 26.20 ± 2.25 | 0.004 |
| C22:4 n-6 | 29.56 ± 2.33 | 32.81 ± 1.64 | 0.144 |
| C22:5 n-3 | 64.30 ± 3.61 | 67.75 ± 2.77 | 0.015 |
| C22:6 n-3 | 50.03 ± 4.65 | 53.60 ± 4.67 | 0.324 |
| Regulated Pathway | Number of Genes | p-Value | Involved Genes (Fold Change) 1 |
|---|---|---|---|
| Integrin Signaling | 13 | 0.001 | ARF5 (+1.34), ARHGAP5 (−1.34), ARPC5L (+1.36), BRAF (−1.37), FYN (−1.83), ILK (+1.41), ITGA1 (−1.52), ITGA5 (+1.61), MYLK2 (+1.27), PIK3C2A (−1.37), PIK3CB (−1.49), PPP1CB (−1.36), TSPAN6 (−1.48) |
| Ephrin A Signaling | 6 | 0.002 | ADAM10 (−1.38), EPHA5 (+1.3), FYN (−1.83), PIK3C2A (−1.37), PIK3CB (−1.49), VAV3 (−1.55) |
| Adipogenesis pathway | 9 | 0.002 | BMPR2 (−1.49), CLOCK (−1.65), DDIT3 (+1.35), FZD5 (−1.35), GTF2H5 (−1.4), HDAC2 (−1.31), KLF3 (−1.34), SIRT1 (−1.42), TXNIP (−1.75) |
| Insulin Receptor Signaling | 8 | 0.010 | CBL (−1.33), FYN (−1.83), INSR (−1.42), PIK3C2A (−1.37), PIK3CB (−1.49), PPP1CB (−1.36), PRKAG2 (−1.33), PTPRF (+1.27) |
| Themes/Biofunctions | p-Value | Involved Genes (Fold Change) 1 |
|---|---|---|
| Carbohydrate Metabolism | ||
| Uptake of d-glucose | <0.001 | CBL (−1.33), CYLD (−1.51), DPP4 (−1.45), EGLN3 (+1.32), GNAS (+1.65), HGF (−1.42), IDH1 (−1.38), INSR (−1.42), MYO1C (+1.36), PDK2 (+1.54), PIK3C2A (−1.37), PIK3CB (−1.49), PPM1A (−1.34), PTPRF (+1.27), SIRT1 (−1.42), TXNIP (−1.75) |
| Quantity of glycogen | 0.001 | GNAS (+1.65), IL6ST (−1.38), INSR (−1.42), LIFR (−1.34), NR1H4 (−1.34), RPS6KA3 (−1.32), SC5D (−1.35), XPA (−1.39) |
| Uptake of carbohydrate | 0.002 | CBL (−1.33), CYLD (−1.51), DPP4 (−1.45), EGLN3 (+1.32), GNAS (+1.65), HGF (−1.42), IDH1 (−1.38), INSR (−1.42), MYO1C (+1.36), NR1H4 (−1.34), PDK2 (+1.54), PIK3C2A (−1.37), PIK3CB (−1.49), PPM1A (−1.34), PTPRF (+1.27), SIRT1 (−1.42), TXNIP (−1.75) |
| Oxidation of carbohydrate | 0.003 | ESRRG (+1.33), INSR (−1.42), PDK2 (+1.54), PNPLA8 (−1.34), SIRT1 (−1.42) |
| Quantity of carbohydrate | 0.010 | ESR1 (+1.36), FOXA1 (+1.32), GNAS (+1.65), GPR39 (+1.39), HGF (−1.42), IL6ST (−1.38), INSR (−1.42), ITPR2 (−1.32), KDM3A (−1.46), LIFR (−1.34), NR1H4 (−1.34), PNPLA8 (−1.34), PSEN2 (−1.39), RPS6KA3 (−1.32), SC5D (−1.35), SGMS2 (−1.50), SIRT1 (−1.42), SLC25A13 (+1.30), SLC3A2 (+1.39), STEAP3 (+1.37), TXNIP (−1.75), VPS13C (−1.33), XPA (−1.39) |
| Disposal of d-glucose | 0.010 | INSR (−1.42), NR1H4 (−1.34), PTPRF (+1.27) |
| Oxidation of d-glucose | 0.012 | ESRRG (+1.33), INSR (−1.42), PDK2 (+1.54), PNPLA8 (−1.34) |
| Phosphorylation of phosphatidylinositol | 0.015 | FAM126A (−1.37), PIK3C2A (−1.37), PIK3CB (−1.49) |
| Import of carbohydrate | 0.019 | B4GALT1 (+1.35), ESR1 (+1.36), INSR (−1.42), PRKAG2 (−1.33), TXNIP (−1.75) |
| Lipid Metabolism | ||
| Synthesis of steroid | 0.004 | ACAT1 (−1.31), BMPR2 (−1.49), ESR1 (+1.36), FOXA1 (+1.32), HGF (−1.42), NR1H4 (−1.34), PDE8A (−1.31), PRKAG2 (−1.33), SIRT1 (−1.42), SLC9A3R2 (+1.33), TLR3 (−1.34), TLR4 (−1.40), TRERF1 (−1.37) |
| Steroidogenesis of cells | 0.004 | SLC9A3R2 (+1.33), TLR3 (−1.34), TLR4 (−1.40) |
| Synthesis of thromboxane | 0.005 | NTN1 (+1.32), PIK3CB (−1.49), PNPLA8 (−1.34) |
| Concentration of fatty acid | 0.006 | CBL (−1.33), GNAS (+1.65), IDH1 (−1.38), INSR (−1.42), ITGA1 (−1.43), KDM3A (−1.46), NR1H4 (−1.34), NTN1 (+1.32), PNPLA8 (−1.34), SIRT1 (−1.42), SLC25A13 (+1.30), SNRK (−1.35), TXNIP (−1.75), XPA (−1.39) |
| Concentration of acylglycerol | 0.016 | CBL (−1.33), FOXA1 (+1.32), GNAS (+1.65), HGF (−1.42), IDH1 (−1.38), INSR (−1.42), ITGA1 (−1.43), KDM3A (−1.46), NR1H4 (−1.34), PDK2 (+1.54), PNPLA8 (−1.34), SGMS2 (−1.5), SIRT1 (−1.42), SLC25A13 (+1.30), SNRK (−1.35), TXNIP (−1.75) |
| Concentration of triacylglycerol | 0.017 | CBL (−1.33), FOXA1 (+1.32), GNAS (+1.65), HGF (−1.42), IDH1 (−1.38), INSR (−1.42), ITGA1 (−1.43), KDM3A (−1.46), NR1H4 (−1.34), PDK2 (+1.54), PNPLA8 (−1.34), SIRT1 (−1.42), SLC25A13 (+1.30), SNRK (−1.35), TXNIP (−1.75) |
| Amino Acid Metabolism | ||
| Transport of neutral amino acid | 0.013 | SLC1A4 (−1.66), SLC3A2 (+1.39), SLC7A9 (+1.50) |
| Transcript | Microarray | RT-qPCR | Correlation | |||||
|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Probe-Set ID | FC 1 | p-Value | q-Value | FC 1 | p-Value | Coefficient | p-Value |
| ITGA5 | SNOWBALL_006991 | +1.61 | <0.001 | 0.028 | +1.82 | 0.011 | 0.94 | <0.001 |
| NR1H4 | SNOWBALL_007505 | −1.34 | 0.002 | 0.181 | −1.29 | 0.048 | 0.69 | 0.013 |
| SLC1A4 | SNOWBALL_005484 | −1.66 | <0.001 | 0.022 | −1.69 | 0.003 | 0.91 | <0.001 |
| SLC7A9 | SNOWBALL_026778 | +1.87 | <0.001 | 0.010 | +1.87 | 0.017 | 0.85 | 0.001 |
| SQLE | SNOWBALL_000764 | −1.82 | <0.001 | 0.004 | −1.89 | 0.005 | 0.91 | <0.001 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyer, H.; Oster, M.; Magowan, E.; Dannenberger, D.; Ponsuksili, S.; Wimmers, K. Strategies towards Improved Feed Efficiency in Pigs Comprise Molecular Shifts in Hepatic Lipid and Carbohydrate Metabolism. Int. J. Mol. Sci. 2017, 18, 1674. https://doi.org/10.3390/ijms18081674
Reyer H, Oster M, Magowan E, Dannenberger D, Ponsuksili S, Wimmers K. Strategies towards Improved Feed Efficiency in Pigs Comprise Molecular Shifts in Hepatic Lipid and Carbohydrate Metabolism. International Journal of Molecular Sciences. 2017; 18(8):1674. https://doi.org/10.3390/ijms18081674
Chicago/Turabian StyleReyer, Henry, Michael Oster, Elizabeth Magowan, Dirk Dannenberger, Siriluck Ponsuksili, and Klaus Wimmers. 2017. "Strategies towards Improved Feed Efficiency in Pigs Comprise Molecular Shifts in Hepatic Lipid and Carbohydrate Metabolism" International Journal of Molecular Sciences 18, no. 8: 1674. https://doi.org/10.3390/ijms18081674
APA StyleReyer, H., Oster, M., Magowan, E., Dannenberger, D., Ponsuksili, S., & Wimmers, K. (2017). Strategies towards Improved Feed Efficiency in Pigs Comprise Molecular Shifts in Hepatic Lipid and Carbohydrate Metabolism. International Journal of Molecular Sciences, 18(8), 1674. https://doi.org/10.3390/ijms18081674