Improvement of Verticillium Wilt Resistance by Applying Arbuscular Mycorrhizal Fungi to a Cotton Variety with High Symbiotic Efficiency under Field Conditions
Abstract
:1. Introduction
2. Results
2.1. Symbiotic Efficiency of Seventeen Cotton Varieties Inoculated by Arbuscular Mycorrhizal Fungi (AMF) R. irregularis
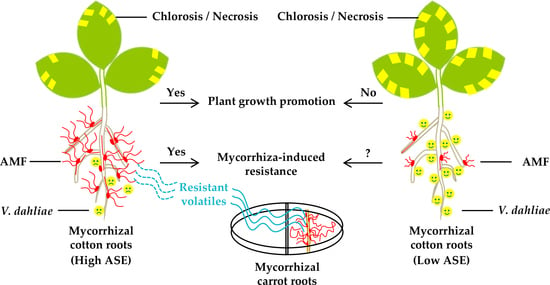
2.2. Effect of Plant Growth Promotion (PGP) among Cotton Varieties with Different Symbiotic Efficiency of AMF (SEA)
2.3. Field Evaluation of PGP of Lumian 1 by Applying for AMF
2.4. Field Performance of Lumian 1 Inoculated with AMF for Verticillium Wilt Resistance
2.5. Effect of Mycorrhizal Colonization on Cotton Resistance-Related Genes
2.6. In Vitro Antifungal Activity Assay of AMF Symbionts
3. Discussion
4. Materials and Methods
4.1. AMF Inoculum Preparation
4.2. Evaluation of SEA among 17 Cotton Cultivars
4.3. Experimental Design for Testing the PGP and Wilt Disease Resistance
4.4. RNA Extraction and Reverse Transcription Quantitative PCR Analysis
4.5. Quantitative Detection of V. dahliae Biomass in Cotton
4.6. Antimicrobial Activities of AMF against Phytopathogenic Fungi In Vitro Systems
4.7. Measurement of Pi Content
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AMF | Arbuscular mycorrhizal fungi |
| PGP | Plant growth promotion |
| SEA | Symbiotic efficiency of AMF |
| MIR | Mycorrhiza-induced resistance |
| SAR | Systemic acquired resistance |
| ISR | Induced systemic resistance |
| MAMP | Microbe-associated molecular patterns |
| PTI | MAMP-triggered immunity |
| SA | Salicylic acid |
| ABA | Abscisic acid |
| DI | Disease index |
| dpi | Days post inoculation |
| JA | Jasmonic acid |
| PR | Pathogenesis-related proteins |
References
- Zhao, Y.; Wang, H.; Chen, W.; Zhao, P.; Gong, H.; Sang, X.; Cui, Y. Regional association analysis-based fine mapping of three clustered QTL for Verticillium wilt resistance in cotton (G. hirsutum L.). BMC Genom. 2017, 18, 661. [Google Scholar] [CrossRef] [PubMed]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Meschke, H.; Walter, S.; Schrempf, H. Characterization and localization of prodiginines from Streptomyces lividans suppressing Verticillium dahliae in the absence or presence of Arabidopsis thaliana. Environ. Microbiol. 2012, 14, 940–952. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liang, C.; Wu, S.; Zhang, X.; Tang, J.; Jian, G.; Jiao, G.; Li, F.; Cui, C. Significant improvement of cotton Verticillium wilt resistance by manipulating the expression of Gastrodia antifungal proteins. Mol. Plant 2016, 9, 1436–1439. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Fan, R.; Dong, H.; Shang, W.; Xu, X.; Zhu, H.; Yang, J.; Hu, X. Threshold microsclerotial inoculum for cotton Verticillium wilt determined through wet-sieving and real-time quantitative PCR. Phytopathology 2015, 105, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Yadeta, K.A.; Hanemian, M.; Smit, P.; Hiemstra, J.A.; Pereira, A.; Marco, Y.; Thomma, B.P. The Arabidopsis thaliana DNA-binding protein AHL19 mediates Verticillium wilt resistance. Mol. Plant Microbe Interact. 2011, 24, 1582–1591. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Fang, H.; Sanogo, S.; Hughs, S.E.; Jones, D.C.; Zhang, J. Evaluation of Verticillium wilt resistance in commercial cultivars and advanced breeding lines of cotton. Euphytica 2014, 196, 437–448. [Google Scholar] [CrossRef]
- Miao, W.; Wang, X.; Li, M.; Song, C.; Wang, Y.; Hu, D.; Wang, J. Genetic transformation of cotton with a harpin-encoding gene hpaXoo confers an enhanced defense response against different pathogens through a priming mechanism. BMC Plant Biol. 2010, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.F.; Ding, Z.G.; Ma, Q.; Zhang, G.R.; Zhang, S.L.; Li, Z.K.; Wu, L.Q.; Zhang, G.Y.; Ma, Z.Y. Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genom. 2013, 14, 637. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.F.; Rong, W.; Yang, J.; Li, Z.K.; Wu, L.Q.; Zhang, G.Y.; Ma, Z.Y. Histochemical analyses reveal that stronger intrinsic defenses in Gossypium barbadense than in G. hirsutum are associated with resistance to Verticillium dahliae. Mol. Plant Microbe Interact. 2017, 30, 984–996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.W.; Zhang, H.C.; Qi, F.J.; Jian, G.L. Generation of transcriptome profiling and gene functional analysis in Gossypium hirsutum upon Verticillium dahliae infection. Biochem. Biophys. Res. Commun. 2016, 473, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.N.; Yuan, Y.L.; Yang, C.; Geng, S.P.; Sun, Q.; Long, L.; Cai, C.W.; Chu, Z.Y.; Liu, X.; Wang, G.H.; et al. Characterization, expression, and functional analysis of a novel NAC gene associated with resistance to Verticillium Wilt and abiotic stress in Cotton. G3 2016, 6, 3951–3961. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Mu, X.Y.; Liu, C.; Cai, J.H.; Shi, K.; Zhu, W.J.; Yang, Q. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integr. Plant Biol. 2015, 57, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.W.; Ling, X.T.; Chen, T.Z.; Cai, L.W.; Liu, T.L.; Wang, J.Y.; Fan, X.H.; Ren, Y.Z.; Yuan, H.B.; Zhu, W.; et al. A cotton Gbvdr5 gene encoding a leucine-rich-repeat receptor-like protein confers resistance to Verticillium dahliae in transgenic Arabidopsis and upland cotton. Plant Mol. Biol. Report. 2015, 33, 987–1001. [Google Scholar] [CrossRef]
- Chen, J.Y.; Xiao, H.L.; Gui, Y.J.; Zhang, D.D.; Li, L.; Bao, Y.M.; Dai, X.F. Characterization of the Verticillium dahliae exoproteome involves in pathogenicity from cotton-containing medium. Front. Microbiol. 2016, 7, 1709. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Li, N.Y.; Ma, X.F.; Gupta, V.K.; Zhang, D.D.; Li, T.G.; Dai, X.F. The ectopic overexpression of the cotton Ve1 and Ve2-homolog sequences leads to resistance response to Verticillium wilt in Arabidopsis. Front. Plant Sci. 2017, 8, 844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Jin, Y.; Zhao, J.H.; Gao, F.; Zhou, B.J.; Fang, Y.Y.; Guo, H.S. Host-induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae. Mol. Plant 2016, 9, 939–942. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.C.; Martinez-Medina, A.; Lopez-Raez, J.A.; Pozo, M.J. Mycorrhiza-induced resistance and priming of plant defenses. J. Chem. Ecol. 2012, 38, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.Y.; Ye, M.; Li, C.Y.; Wang, R.L.; Wei, X.C.; Luo, S.M.; Zeng, R.S. Priming of anti-herbivore defense in tomato by arbuscular mycorrhizal fungus and involvement of the jasmonate pathway. J. Chem. Ecol. 2013, 39, 1036–1044. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Baccelli, I.; Luna, E.; Flors, V. Defense priming: An adaptive part of induced resistance. Annu. Rev. Plant Biol. 2017, 68, 485–512. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.Y.; Chen, D.M.; Lu, K.; Sun, Z.X.; Zeng, R.S. Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front. Plant Sci. 2015, 6, 786. [Google Scholar] [CrossRef] [PubMed]
- Cameron, D.D.; Neal, A.L.; van Wees, S.C.; Ton, J. Mycorrhiza-induced resistance: More than the sum of its parts? Trends Plant Sci. 2013, 18, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Nadal, M.; Sawers, R.; Naseem, S.; Bassin, B.; Kulicke, C.; Sharman, A.; An, G.; An, K.; Ahern, K.R.; Romag, A.; et al. An N-acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize. Nat. Plants 2017, 3, 17073. [Google Scholar] [CrossRef] [PubMed]
- Genre, A.; Chabaud, M.; Balzergue, C.; Puech-Pagès, V.; Novero, M.; Rey, T.; Fournier, J.; Rochange, S.; Bécard, G.; Bonfante, P.; et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013, 198, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, J.M. Plant immunity triggered by microbial molecular signatures. Mol. Plant 2010, 3, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; Bruce, T.J.; Roberts, M.R.; Flors, V.; Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 2012, 158, 844–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blilou, I.; Ocampo, J.A.; García-Garrido, J.M. Resistance of pea roots to endomycorrhizal fungus or Rhizobium correlates with enhanced levels of endogenous salicylic acid. J. Exp. Bot. 1999, 50, 1663–1668. [Google Scholar] [CrossRef]
- De Jonge, R.; Bolton, M.D.; Thomma, B.P. How filamentous pathogens co-opt plants: The ins and outs of fungal effectors. Curr. Opin. Plant Biol. 2011, 14, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Ton, J.; Flors, V.; Mauch-Mani, B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Van Wees, S.C.; Van der Ent, S.; Pieterse, C.M. Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 2008, 11, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Cotton, T.A.; Dumbrell, A.J.; Helgason, T. What goes in must come out: Testing for biases in molecular analysis of arbuscular mycorrhizal fungal communities. PLoS ONE 2014, 9, e109234. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.N.; Wang, W.X.; Xie, Q.J.; Liu, N.; Liu, L.X.; Wang, D.P.; Zhang, X.W.; Yang, C.; Chen, X.Y.; Tang, D.Z.; et al. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 2017, 356, 1172–1175. [Google Scholar] [CrossRef] [PubMed]
- Luginbuehl, L.H.; Menard, G.N.; Kurup, S.; Van Erp, H.; Radhakrishnan, G.V.; Breakspear, A.; Oldroyd, G.E.D.; Eastmond, P.J. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 2017, 356, 1175–1178. [Google Scholar] [CrossRef] [PubMed]
- Parniske, M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008, 6, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Garg, N.; Chandel, S. Arbuscular mycorrhizal networks: Process and functions. A review. Agron. Sustain. Dev. 2010, 30, 581–599. [Google Scholar] [CrossRef]
- Smith, S.E.; Smith, F.A. Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 2011, 62, 227–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Fattah, G.M.; El-Haddad, S.A.; Hafez, E.E.; Rashad, Y.M. Induction of defense responses in common bean plants by arbuscular mycorrhizal fungi. Microbiol. Res. 2011, 166, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.J. Effect of vesicular-arbuscular mycorrhizal fungi on Verticillium wilt of cotton. Mycorrhiza 1995, 5, 293–297. [Google Scholar] [CrossRef]
- Eke, P.; Chatue, G.C.; Wakam, L.N.; Kouipou, R.M.T.; Fokou, P.V.T.; Boyom, F.F. Mycorrhiza consortia suppress the fusarium root rot (Fusarium solani f. sp. Phaseoli) in common bean (Phaseolus vulgaris L.). Biol. Control 2016, 103, 240–250. [Google Scholar] [CrossRef]
- Sukhada, M.; Manjula, R.; Rawal, R.D. Evaluation of arbuscular mycorrhiza and other biocontrol agents against Phytophthora parasitica var. nicotianae infecting papaya (Carica papaya cv. Surya) and enumeration of pathogen population using immunotechniques. Biol. Control 2011, 58, 22–29. [Google Scholar] [CrossRef]
- Spagnoletti, F.N.; Balestrasse, K.; Lavado, R.S.; Giacometti, R. Arbuscular mycorrhiza detoxifying response against arsenic and pathogenic fungus in soybean. Ecotoxicol. Environ. Saf. 2016, 133, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Franken, P. Comparison of systemic and local interactions between the arbuscular mycorrhizal fungus Funneliformis mosseae and the root pathogen Aphanomyces euteiches in Medicago truncatula. Mycorrhiza 2014, 24, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Cely, M.V.; de Oliveira, A.G.; de Freitas, V.F.; de Luca, M.B.; Barazetti, A.R.; Dos Santos, I.M.; Gionco, B.; Garcia, G.V.; Prete, C.E.; Andrade, G. Inoculant of arbuscular mycorrhizal fungi (Rhizophagus clarus) increase yield of soybean and cotton under field conditions. Front. Microbiol. 2016, 7, 720. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.X.; Wang, D.B.; Zhou, Q. Colonization and persistence of a plant growth-promoting bacterium Pseudomonas fluorescens strain CS85, on roots of cotton seedlings. Can. J. Microbiol. 2004, 50, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Uehara, Y.; Berberich, T.; Ito, A.; Saitoh, H.; Miyazaki, A.; Terauchi, R.; Kusano, T. A subset of hypersensitive response marker genes, including HSR203J, is the downstream target of a spermine signal transduction pathway in tobacco. Plant J. 2004, 40, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Klessig, D.F.; Nürnberger, T. A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene HIN1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. Plant Cell 2001, 13, 1079–1093. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Park, Y.J.; Seo, P.J.; Kim, J.H.; Sim, H.J.; Kim, S.G.; Park, C.M. Systemic immunity requires SnRK2.8-mediated nuclear import of NPR1 in Arabidopsis. Plant Cell 2015, 27, 3425–3438. [Google Scholar] [CrossRef] [PubMed]
- Colditz, F.; Niehaus, K.; Krajinski, F. Silencing of PR-10-like proteins in Medicago truncatula results in an antagonistic induction of other PR proteins and in an increased tolerance upon infection with the oomycete Aphanomyces euteiches. Planta 2007, 226, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhu, L.F.; Tu, L.L.; Liu, L.L.; Yuan, D.J.; Jin, L.; Long, L.; Zhang, X.L. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 2011, 62, 5607–5621. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.F.; Jin, L.; Miao, Y.H.; He, X.; Hu, Q.; Guo, K.; Zhu, L.F.; Zhang, X.L. An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 2016, 91, 305–318. [Google Scholar] [CrossRef] [PubMed]
- López-Ráez, J.A.; Verhage, A.; Fernández, I.; García, J.M.; Azcón-Aguilar, C.; Flors, V.; Pozo, M.J. Hormonal and transcriptional profiles highlight common and differential host responses to arbuscular mycorrhizal fungi and the regulation of the oxylipin pathway. J. Exp. Bot. 2010, 61, 2589–2601. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Shen, Y.; Hao, J.; Wu, J.; Ke, L.; Wu, C.; Huang, K.; Luo, B.; Xu, M.; Cheng, X.; et al. Acyl-CoA N-acyltransferase influences fertility by regulating lipid metabolism and jasmonic acid biogenesis in cotton. Sci. Rep. 2015, 5, 11790. [Google Scholar] [CrossRef] [PubMed]
- Volpe, V.; Giovannetti, M.; Sun, X.G.; Fiorilli, V.; Bonfante, P. The phosphate transporters LjPT4 and MtPT4 mediate early root responses to phosphate status in non mycorrhizal roots. Plant Cell Environ. 2016, 39, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Lin, H.; Peng, X.; Xu, C.; Sun, Z.; Jiang, K.; Huang, A.; Wu, X.; Tang, N.; Salvioli, A.; et al. Arbuscular mycorrhizal symbiosis requires a phosphate transceptor in the Gigaspora margarita fungal symbiont. Mol. Plant 2016, 9, 1583–1608. [Google Scholar] [CrossRef] [PubMed]
- Bender, S.F.; Conen, F.; Van der Heijden, M.G. Mycorrhizal effects on nutrient cycling, nutrient leaching and N2O production in experimental grassland. Soil Biol. Biochem. 2015, 80, 283–292. [Google Scholar] [CrossRef]
- Calabrese, S.; Pérez-Tienda, J.; Ellerbeck, M.; Arnould, C.; Chatagnier, O.; Boller, T.; Schüßler, A.; Brachmann, A.; Wipf, D.; Ferrol, N.; et al. GintAMT3-a low-Affinity Ammonium Transporter of the Arbuscular Mycorrhizal Rhizophagus irregularis. Front. Plant Sci. 2016, 7, 679. [Google Scholar] [CrossRef] [PubMed]
- Garcia, K.; Chasman, D.; Roy, S.; Ané, J. Physiological responses and gene co-expression network of mycorrhizal roots under K+ deprivation. Plant Physiol. 2017, 173, 1811–1823. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Miller, R.M. Carbon cycling by arbuscular mycorrhizal fungi in soil–plant systems. Trends Plant Sci. 2003, 8, 407–409. [Google Scholar] [CrossRef]
- Verbruggen, E.; Heijden, M.G.; Rillig, M.C.; Kiers, E.T. Mycorrhizal fungal establishment in agricultural soils: Factors determining inoculation success. New Phytol. 2013, 197, 1104–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, A.M.; Croll, D.; Sanders, I.R. Genetic variability in a population of arbuscular mycorrhizal fungi causes variation in plant growth. Ecol. Lett. 2006, 9, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Munkvold, L.; Kjøller, R.; Vestberg, M.; Rosendahl, S.; Jakobsen, I. High functional diversity within species of arbuscular mycorrhizal fungi. New Phytol. 2004, 164, 357–364. [Google Scholar] [CrossRef]
- Colard, A.; Angelard, C.; Sanders, I.R. Genetic exchange in an arbuscular mycorrhizal fungus results in increased rice growth and altered mycorrhiza-specific gene transcription. Appl. Environ. Microbiol. 2011, 77, 6510–6515. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M. Effect of indigenous arbuscular mycorrhizal fungi combined with manure on the change in concentration some mineral elements in cotton (Gossypium hirsutum L.). J. Plant Nutr. 2017, 40, 2862–2871. [Google Scholar] [CrossRef]
- Eason, W.R.; Webb, K.J.; Michaelson-Yeates, T.P.T.; Abberton, M.T.; Griffith, G.W.; Culshaw, C.M.; Hooker, J.E.; Dhanoa, M.S. Effect of genotype of Trifolium repens on mycorrhizal symbiosis with Glomus mosseae. J. Agric. Sci. 2001, 137, 27–36. [Google Scholar] [CrossRef]
- MacLean, A.M.; Bravo, A.; Harrison, M.J. Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. Plant Cell 2017, 29, 2319–2335. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, V.K.; Jain, A.; Poling, M.D.; Lewis, A.J.; Raghothama, K.G.; Smith, A.P. Arabidopsis Pht1; 5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 2011, 156, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, H.; Michalska, K.; Sikorski, M.; Jaskolski, M.; Raghothama, K.G.; Smith, A.P. Structural and functional aspects of PR-10 proteins. FEBS J. 2013, 280, 1169–1199. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.R.; Chen, Z.Y.; Brown, R.L.; Bhatnagar, D. Expression and functional characterization of two pathogenesis-related protein 10 genes from Zea mays. J. Plant Physiol. 2010, 167, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Constabel, C.P.; Bertrand, C.; Brisson, N. Transgenic potato plants overexpressing the pathogenesis-related STH-2 gene show unaltered susceptibility to Phytophthora infestans and potato virus X. Plant Mol. Biol. 1993, 22, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.S.; Huang, J.C.; Hu, J.H. Characterization of two subclasses of PR-10 transcripts in lily anthers and induction of their genes through separate signal transduction pathways. Plant Mol. Biol. 1999, 40, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.R. Analysis of PR Protein Family and Functional Study of PR10 and PR17 in Cotton. Master’s Thesis, Agricultural University of Hebei, Baoding, China, 2015. [Google Scholar]
- Liang, S. Isolation and Characterization of Cotton PR10-Like Gene Responsive to Infection by Verticillium dahliae. Master’s Thesis, Hebei University, Baoding, China, 2009. [Google Scholar]
- Zhang, G.; Raza, W.; Wang, X.; Ran, W.; Shen, Q. Systemic modification of cotton root exudates induced by arbuscular mycorrhizal fungi and Bacillus vallismortis HJ-5 and their effects on Verticillium wilt disease. Appl. Soil Ecol. 2012, 61, 85–91. [Google Scholar] [CrossRef]
- Suh, H.W.; Crawford, D.L.; Korus, R.A.; Shetty, K. Production of antifungal metabolites by the ectomycorrhizal fungus Pisolithus tinctorius strain SMF. J. Ind. Microbiol. 1991, 8, 29–35. [Google Scholar] [CrossRef]
- Grunewaldt-Stöcker, G.; von den Berg, C.; Knopp, J.; von Alten, H. Interactions of ericoid mycorrhizal fungi and root pathogens in Rhododendron: In vitro tests with plantlets in sterile liquid culture. Plant Root 2013, 7, 33–48. [Google Scholar] [CrossRef]
- Hage-Ahmed, K.; Moyses, A.; Voglgruber, A.; Hadacek, F.; Steinkellner, S. Alterations in root exudation of intercropped tomato mediated by the arbuscular mycorrhizal fungus Glomus mosseae and the soilborne pathogen Fusarium oxysporum f.sp. lycopersici. J. Phytopathol. 2013, 161, 763–773. [Google Scholar] [CrossRef]
- St-Arnaud, M.; Hamel, C.; Vimard, B.; Caron, M.; Fortin, J.A. Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol. Res. 1996, 100, 328–332. [Google Scholar] [CrossRef]
- Phillips, J.M.; Hayman, D.S. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 1970, 55, 158–161. [Google Scholar] [CrossRef]
- Mo, H.J.; Sun, Y.X.; Zhu, X.L.; Wang, X.F.; Zhang, Y.; Yang, J.; Yan, G.J.; Ma, Z.Y. Cotton S-adenosylmethionine decarboxylase-mediated spermine biosynthesis is required for salicylic acid- and leucine-correlated signaling in the defense response to Verticillium dahliae. Planta 2016, 243, 1023–1039. [Google Scholar] [CrossRef] [PubMed]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Gkizi, D.; Lehmann, S.; L’Haridon, F.; Serrano, M.; Paplomatas, E.J.; Métraux, J.P.; Tjamos, S.E. The innate immune signaling system as a regulator of disease resistance and induced systemic resistance activity against Verticillium dahliae. Mol. Plant Microbe Interact. 2016, 29, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Nanamori, M.; Shinano, T.; Wasaki, J.; Yamamura, T.; Rao, I.M.; Osaki, M. Low phosphorus tolerance mechanisms: Phosphorus recycling and photosynthate partitioning in the tropical forage grass, Brachiaria hybrid cultivar Mulato compared with rice. Plant Cell Physiol. 2004, 45, 460–469. [Google Scholar] [CrossRef] [PubMed]








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Gao, X.; Ren, Y.; Ding, X.; Qiu, J.; Li, N.; Zeng, F.; Chu, Z. Improvement of Verticillium Wilt Resistance by Applying Arbuscular Mycorrhizal Fungi to a Cotton Variety with High Symbiotic Efficiency under Field Conditions. Int. J. Mol. Sci. 2018, 19, 241. https://doi.org/10.3390/ijms19010241
Zhang Q, Gao X, Ren Y, Ding X, Qiu J, Li N, Zeng F, Chu Z. Improvement of Verticillium Wilt Resistance by Applying Arbuscular Mycorrhizal Fungi to a Cotton Variety with High Symbiotic Efficiency under Field Conditions. International Journal of Molecular Sciences. 2018; 19(1):241. https://doi.org/10.3390/ijms19010241
Chicago/Turabian StyleZhang, Qiang, Xinpeng Gao, Yanyun Ren, Xinhua Ding, Jiajia Qiu, Ning Li, Fanchang Zeng, and Zhaohui Chu. 2018. "Improvement of Verticillium Wilt Resistance by Applying Arbuscular Mycorrhizal Fungi to a Cotton Variety with High Symbiotic Efficiency under Field Conditions" International Journal of Molecular Sciences 19, no. 1: 241. https://doi.org/10.3390/ijms19010241
APA StyleZhang, Q., Gao, X., Ren, Y., Ding, X., Qiu, J., Li, N., Zeng, F., & Chu, Z. (2018). Improvement of Verticillium Wilt Resistance by Applying Arbuscular Mycorrhizal Fungi to a Cotton Variety with High Symbiotic Efficiency under Field Conditions. International Journal of Molecular Sciences, 19(1), 241. https://doi.org/10.3390/ijms19010241