Identification of Host Defense-Related Proteins Using Label-Free Quantitative Proteomic Analysis of Milk Whey from Cows with Staphylococcus aureus Subclinical Mastitis
Abstract
:1. Introduction
2. Results
3. Discussion
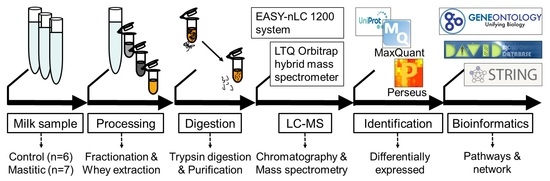
4. Methods
4.1. Sample Collection
4.2. SCC and Microbiological Examination
4.3. Separation of Whey Proteins
4.4. In-Solution Trypsin Digestion of Whey Proteins
4.5. Liquid Chromatography and Tandem Mass Spectrometry (LC-MS/MS)
4.6. Quantitative Proteomic Data Analysis
4.6.1. Protein Identification and Label-Free Quantification
4.6.2. Statistical Analysis
4.6.3. Gene Ontology (GO) Enrichment Analysis
4.6.4. Pathway and Network Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hogeveen, H.; Huijps, K.; Lam, T.J. Economic aspects of mastitis: New developments. N. Z. Vet. J. 2011, 59, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Halasa, T.; Huijps, K.; Osteras, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Seegers, H.; Fourichon, C.; Beaudeau, F. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet. Res. 2003, 34, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Barkema, H.W.; Schukken, Y.H.; Zadoks, R.N. Invited Review: The role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitis. J. Dairy Sci. 2006, 89, 1877–1895. [Google Scholar] [CrossRef]
- Olde Riekerink, R.G.; Barkema, H.W.; Scholl, D.T.; Poole, D.E.; Kelton, D.F. Management practices associated with the bulk-milk prevalence of Staphylococcus aureus in Canadian dairy farms. Prev. Vet. Med. 2010, 97, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Ambatipudi, K. Challenges and opportunities of bovine milk analysis by mass spectrometry. Clin. Proteom. 2016, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- Viguier, C.; Arora, S.; Gilmartin, N.; Welbeck, K.; O’Kennedy, R. Mastitis detection: Current trends and future perspectives. Trends Biotechnol. 2009, 27, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Ceciliani, F.; Eckersall, D.; Burchmore, R.; Lecchi, C. Proteomics in veterinary medicine: Applications and trends in disease pathogenesis and diagnostics. Vet. Pathol. 2014, 51, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Wellnitz, O.; Bruckmaier, R.M. The innate immune response of the bovine mammary gland to bacterial infection. Vet. J. 2012, 192, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Souza, F.; Ramos Sanchez, E.; Heinemann, M.; Gidlund, M.; Reis, L.; Blagitz, M.; Della Libera, A.; Cerqueira, M. The innate immunity in bovine mastitis: The role of pattern-recognition receptors. Am. J. Immunol. 2012, 8, 166–178. [Google Scholar] [CrossRef]
- Rainard, P.; Riollet, C. Innate immunity of the bovine mammary gland. Vet. Res. 2006, 37, 369–400. [Google Scholar] [CrossRef] [PubMed]
- Boehmer, J.L. Proteomic analyses of host and pathogen responses during bovine mastitis. J. Mammary Gland Biol. Neoplas. 2011, 16, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Smolenski, G.A.; Broadhurst, M.K.; Stelwagen, K.; Haigh, B.J.; Wheeler, T.T. Host defence related responses in bovine milk during an experimentally induced Streptococcus uberis infection. Proteome Sci. 2014, 12, 19. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.F.; Bronzo, V.; Puggioni, G.M.G.; Cacciotto, C.; Tedde, V.; Pagnozzi, D.; Locatelli, C.; Casula, A.; Curone, G.; Uzzau, S.; et al. Relationship between milk cathelicidin abundance and microbiologic culture in clinical mastitis. J. Dairy Sci. 2017, 100, 2944–2953. [Google Scholar] [CrossRef] [PubMed]
- Boehmer, J.L.; DeGrasse, J.A.; McFarland, M.A.; Tall, E.A.; Shefcheck, K.J.; Ward, J.L.; Bannerman, D.D. The proteomic advantage: Label-free quantification of proteins expressed in bovine milk during experimentally induced coliform mastitis. Vet. Immunol. Immunopathol. 2010, 138, 252–266. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, T.A.; Sacco, R.E.; Nonnecke, B.J.; Lippolis, J.D. Bovine milk proteome: Quantitative changes in normal milk exosomes, milk fat globule membranes and whey proteomes resulting from Staphylococcus aureus mastitis. J. Proteom. 2013, 82, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Atalla, H.; Mallard, B.; Robert, C.; Karrow, N. Changes in Holstein cow milk and serum proteins during intramammary infection with three different strains of Staphylococcus aureus. BMC Vet. Res. 2011, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Larsen, L.B.; Hinz, K.; Jorgensen, A.L.; Moller, H.S.; Wellnitz, O.; Bruckmaier, R.M.; Kelly, A.L. Proteomic and peptidomic study of proteolysis in quarter milk after infusion with lipoteichoic acid from Staphylococcus aureus. J. Dairy Sci. 2010, 93, 5613–5626. [Google Scholar] [CrossRef] [PubMed]
- Bishop, S.C.; Woolliams, J.A. Genomics and disease resistance studies in livestock. Livest. Sci. 2014, 166, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Bannerman, D.D.; Paape, M.J.; Lee, J.W.; Zhao, X.; Hope, J.C.; Rainard, P. Escherichia coli and Staphylococcus aureus elicit differential innate immune responses following intramammary infection. Clin. Diagn. Lab. Immunol. 2004, 11, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Bannerman, D. Pathogen-dependent induction of cytokines and other soluble inflammatory mediators during intramammary infection of dairy cows. J. Anim. Sci. 2009, 87 (Suppl. 13), 10–25. [Google Scholar] [CrossRef] [PubMed]
- Ceciliani, F.; Ceron, J.J.; Eckersall, P.D.; Sauerwein, H. Acute phase proteins in ruminants. J. Proteom. 2012, 75, 4207–4231. [Google Scholar] [CrossRef] [PubMed]
- Whelehan, C.J.; Meade, K.G.; Eckersall, P.D.; Young, F.J.; O’Farrelly, C. Experimental Staphylococcus aureus infection of the mammary gland induces region-specific changes in innate immune gene expression. Vet. Immunol. Immunopathol. 2011, 140, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Schroder, N.W.; Morath, S.; Alexander, C.; Hamann, L.; Hartung, T.; Zahringer, U.; Gobel, U.B.; Weber, J.R.; Schumann, R.R. Lipoteichoic acid (LTA) of Streptococcus pneumoniae and Staphylococcus aureus activates immune cells via Toll-like receptor (TLR)-2, lipopolysaccharide-binding protein (LBP), and CD14, whereas TLR-4 and MD-2 are not involved. J. Biol. Chem. 2003, 278, 15587–15594. [Google Scholar] [CrossRef] [PubMed]
- Kiku, Y.; Ozawa, T.; Kushibiki, S.; Sudo, M.; Kitazaki, K.; Abe, N.; Takahashi, H.; Hayashi, T. Decrease in bovine CD14 positive cells in colostrum is associated with the incidence of mastitis after calving. Vet. Res. Commun. 2010, 34, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Suojala, L.; Orro, T.; Järvinen, H.; Saatsi, J.; Pyörälä, S. Acute phase response in two consecutive experimentally induced E. coli intramammary infections in dairy cows. Acta Vet. Scand. 2008, 50, 18. [Google Scholar] [CrossRef] [PubMed]
- Bannerman, D.D.; Paape, M.J.; Hare, W.R.; Sohn, E.J. Increased levels of LPS-binding protein in bovine blood and milk following bacterial lipopolysaccharide challenge. J. Dairy Sci. 2003, 86, 3128–3137. [Google Scholar] [CrossRef]
- Thomas, F.C. Acute Phase Proteins, Proteomics and Metabolomics in the Diagnosis of Bovine Mastitis. Ph.D. Thesis, University of Glasgow, Glasgow, UK, 2015. [Google Scholar]
- Thomas, F.C.; Waterston, M.; Hastie, P.; Parkin, T.; Haining, H.; Eckersall, P.D. The major acute phase proteins of bovine milk in a commercial dairy herd. BMC Vet. Res. 2015, 11, 207. [Google Scholar] [CrossRef] [PubMed]
- Morimatsu, M.; Syuto, B.; Shimada, N.; Fujinaga, T.; Yamamoto, S.; Saito, M.; Naiki, M. Isolation and characterization of bovine haptoglobin from acute phase sera. J. Biol. Chem. 1991, 266, 11833–11837. [Google Scholar] [PubMed]
- Grönlund, U.; Hultén, C.; Eckersall, P.D.; Hogarth, C.; Waller, K.P. Haptoglobin and serum amyloid A in milk and serum during acute and chronic experimentally induced Staphylococcus aureus mastitis. J. Dairy Res. 2003, 70, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Ezzat Alnakip, M.; Quintela-Baluja, M.; Böhme, K.; Fernández-No, I.; Caamaño-Antelo, S.; Calo-Mata, P.; Barros-Velázquez, J. The immunology of mammary gland of dairy ruminants between healthy and inflammatory conditions. J. Vet. Med. 2014, 2014, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Stelwagen, K.; Carpenter, E.; Haigh, B.; Hodgkinson, A.; Wheeler, T. Immune components of bovine colostrum and milk. J. Anim. Sci. 2009, 87 (Suppl. 13), 3–9. [Google Scholar] [CrossRef]
- Cubeddu, T.; Cacciotto, C.; Pisanu, S.; Tedde, V.; Alberti, A.; Pittau, M.; Dore, S.; Cannas, A.; Uzzau, S.; Rocca, S.; et al. Cathelicidin production and release by mammary epithelial cells during infectious mastitis. Vet. Immunol. Immunopathol. 2017, 189 (Suppl. C), 66–70. [Google Scholar] [CrossRef] [PubMed]
- Chaneton, L.; Tirante, L.; Maito, J.; Chaves, J.; Bussmann, L. Relationship between milk lactoferrin and etiological agent in the mastitic bovine mammary gland. J. Dairy Sci. 2008, 91, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, S.; Kawai, K.; Anri, A.; Nagahata, H. Lactoferrin concentrations in milk from normal and subclinical mastitic cows. J. Vet. Med. Sci. 2003, 65, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Van Altena, S.; de Klerk, B.; Hettinga, K.; van Neerven, R.; Boeren, S.; Savelkoul, H.; Tijhaar, E. A proteomics-based identification of putative biomarkers for disease in bovine milk. Vet. Immunol. Immunopathol. 2016, 174, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Elsik, C.G.; Tellam, R.L.; Worley, K.C. The genome sequence of taurine cattle: A window to ruminant biology and evolution. Science 2009, 324, 522–528. [Google Scholar] [PubMed]
- Zanetti, M. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 2004, 75, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Smolenski, G.; Wieliczko, R.; Pryor, S.; Broadhurst, M.; Wheeler, T.; Haigh, B. The abundance of milk cathelicidin proteins during bovine mastitis. Vet. Immunol. Immunopathol. 2011, 143, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.; Tedde, V.; Dore, S.; Pisanu, S.; Puggioni, G.; Roggio, A.; Pagnozzi, D.; Lollai, S.; Cannas, E.; Uzzau, S. Evaluation of milk cathelicidin for detection of dairy sheep mastitis. J. Dairy Sci. 2016, 99, 6446–6456. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, D.R.; Wang, M.; Liu, L.-H.; Boons, G.-J.; Gupta, D.; Dziarski, R. Peptidoglycan recognition proteins kill bacteria by activating protein-sensing two-component systems. Nat. Med. 2011, 17, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Mudaliar, M.; Tassi, R.; Thomas, F.C.; McNeilly, T.N.; Weidt, S.K.; McLaughlin, M.; Wilson, D.; Burchmore, R.; Herzyk, P.; Eckersall, P.D.; et al. Mastitomics, the integrated omics of bovine milk in an experimental model of Streptococcus uberis mastitis: 2. Label-free relative quantitative proteomics. Mol. Biosyst. 2016, 12, 2748–2761. [Google Scholar] [CrossRef] [PubMed]
- Ibeagha-Awemu, E.M.; Ibeagha, A.E.; Messier, S.; Zhao, X. Proteomics, genomics, and pathway analyses of Escherichia coli and Staphylococcus aureus infected milk whey reveal molecular pathways and networks involved in mastitis. J. Proteome Res. 2010, 9, 4604–4619. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Iwamuro, S. Potential roles of histones in host defense as antimicrobial agents. Infect. Disord. Drug Targets (Former. Curr. Drug Targets-Infect. Disord.) 2008, 8, 195–205. [Google Scholar] [CrossRef]
- Lippolis, J.; Reinhardt, T. Centennial paper: Proteomics in animal science. J. Anim. Sci. 2008, 86, 2430–2441. [Google Scholar] [CrossRef] [PubMed]
- Pisanu, S.; Cubeddu, T.; Pagnozzi, D.; Rocca, S.; Cacciotto, C.; Alberti, A.; Marogna, G.; Uzzau, S.; Addis, M.F. Neutrophil extracellular traps in sheep mastitis. Vet. Res. 2015, 46, 59. [Google Scholar] [CrossRef] [PubMed]
- Lippolis, J.D.; Peterson-Burch, B.D.; Reinhardt, T.A. Differential expression analysis of proteins from neutrophils in the periparturient period and neutrophils from dexamethasone-treated dairy cows. Vet. Immunol. Immunopathol. 2006, 111, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Luo, G.; Zhang, Z.; Wang, X.; Ju, Z.; Qi, C.; Zhang, Y.; Wang, C.; Li, R.; Li, J. iTRAQ-proteomics and bioinformatics analyses of mammary tissue from cows with clinical mastitis due to natural infection with Staphylococci aureus. BMC Genom. 2014, 15, 839. [Google Scholar] [CrossRef] [PubMed]
- National Mastitis Council (US); Research Committee. Microbiological Procedures for the Diagnosis of Bovine Udder Infection, 3rd ed.; National Mastitis Council: Arlington, VA, USA, 1990. [Google Scholar]
- Atalla, H.; Gyles, C.; Jacob, C.L.; Moisan, H.; Malouin, F.; Mallard, B. Characterization of a Staphylococcus aureus small colony variant (SCV) associated with persistent bovine mastitis. Foodborne Pathog. Dis. 2008, 5, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Matic, I.; Hilger, M.; Nagaraj, N.; Selbach, M.; Olsen, J.V.; Mann, M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009, 4, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2013, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Csordas, A.; Sun, Z.; Jarnuczak, A.; Perez-Riverol, Y.; Ternent, T.; Campbell, D.S.; Bernal-Llinares, M.; Okuda, S.; Kawano, S. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2016, 45, D1100–D1106. [Google Scholar] [CrossRef] [PubMed]



| UniProt ID | Protein Name | Log2 Fold Change | p-Value | Unique Peptides Count |
|---|---|---|---|---|
| Q8SPP7 | Peptidoglycan recognition protein 1 | 7.92 (+) | 0.000 | 10 |
| Q2TBU0 | Haptoglobin | 6.45 | 0.002 | 17 |
| Q3ZBX9 | Histone H2A.J | 5.47 | 0.000 | 1 |
| P63258 | Actin, cytoplasmic 2 | 5.46 | 0.002 | 9 |
| E1B6Z6 | Neutrophil gelatinase-associated lipocalin | 5.29 | 0.001 | 13 |
| P10096 | Glyceraldehyde-3-phosphate dehydrogenase | 5.17 | 0.003 | 8 |
| Q1JPB0 | Leukocyte elastase inhibitor (Serpin B1) | 4.31 | 0.003 | 7 |
| P33046 | Cathelicidin-4 (Indolicidin) | 4.29 | 0.002 | 4 |
| G3X807 | Histone H4 | 4.09 | 0.003 | 6 |
| F1MYX5 | Uncharacterized protein | 4.08 | 0.002 | 13 |
| P30922 | Chitinase-3-like protein 1 | 3.97 | 0.01 | 16 |
| G3MXB5 | Uncharacterized protein | 2.70 | 0.002 | 5 |
| Q2TBI0 | Lipopolysaccharide-binding protein | 2.69 | 0.014 | 12 |
| P07688 | Cathepsin B | 2.31 | 0.011 | 9 |
| P24627 | Lactotransferrin (Lactoferrin) | 2.28 | 0.001 | 61 |
| P79345 | Epididymal secretory protein E1 | −0.70 (−) | 0.013 | 9 |
| P08037 | Beta-1,4-galactosyltransferase 1 | −1.08 | 0.011 | 12 |
| Q9TUM6 | Perilipin-2 (Adipophilin) | −1.55 | 0.006 | 18 |
| P11151 | Lipoprotein lipase | −1.57 | 0.007 | 13 |
| Q95122 | Monocyte differentiation antigen CD14 | −1.70 | 0.00 | 9 |
| Q0P569 | Nucleobindin-1 | −1.84 | 0.014 | 15 |
| P80025 | Lactoperoxidase | −1.93 | 0.001 | 31 |
| G5E5H7 | Uncharacterized protein | −2.18 | 0.009 | 3 |
| Q4GZT4 | ATP-binding cassette sub-family G member 2 | −2.33 | 0.004 | 12 |
| Q9XSG3 | Isocitrate dehydrogenase | −2.92 | 0.013 | 10 |
| GO Terms (Biological Processes) * | Gene Name | p-Value |
|---|---|---|
| GO:0042742~defense response to bacterium | Hp, LPO, CATHL4 | 0.0066 |
| GO:0031640~killing of cells of other organism | PGLYRP1, CATHL4 | 0.0076 |
| GO:0071223~cellular response to lipoteichoic acid | LBP, CD14, PGLYRP1 | 0.0091 |
| GO:0034145~positive regulation of Toll-like receptor 4 signaling pathway | LTF, LBP | 0.0136 |
| GO:0006953~acute-phase response | Hp, LBP | 0.027 |
| GO:0019731~antibacterial humoral response | LTF, LPO | 0.0314 |
| GO:0031663~lipopolysaccharide-mediated signaling pathway | LBP, CD14 | 0.0417 |
| GO:0045087~innate immune response | LBP, CD14, PGLYRP1 | 0.0459 |
| GO:0098869~cellular oxidant detoxification | Hp, LPO | 0.0461 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelmegid, S.; Murugaiyan, J.; Abo-Ismail, M.; Caswell, J.L.; Kelton, D.; Kirby, G.M. Identification of Host Defense-Related Proteins Using Label-Free Quantitative Proteomic Analysis of Milk Whey from Cows with Staphylococcus aureus Subclinical Mastitis. Int. J. Mol. Sci. 2018, 19, 78. https://doi.org/10.3390/ijms19010078
Abdelmegid S, Murugaiyan J, Abo-Ismail M, Caswell JL, Kelton D, Kirby GM. Identification of Host Defense-Related Proteins Using Label-Free Quantitative Proteomic Analysis of Milk Whey from Cows with Staphylococcus aureus Subclinical Mastitis. International Journal of Molecular Sciences. 2018; 19(1):78. https://doi.org/10.3390/ijms19010078
Chicago/Turabian StyleAbdelmegid, Shaimaa, Jayaseelan Murugaiyan, Mohamed Abo-Ismail, Jeff L. Caswell, David Kelton, and Gordon M. Kirby. 2018. "Identification of Host Defense-Related Proteins Using Label-Free Quantitative Proteomic Analysis of Milk Whey from Cows with Staphylococcus aureus Subclinical Mastitis" International Journal of Molecular Sciences 19, no. 1: 78. https://doi.org/10.3390/ijms19010078
APA StyleAbdelmegid, S., Murugaiyan, J., Abo-Ismail, M., Caswell, J. L., Kelton, D., & Kirby, G. M. (2018). Identification of Host Defense-Related Proteins Using Label-Free Quantitative Proteomic Analysis of Milk Whey from Cows with Staphylococcus aureus Subclinical Mastitis. International Journal of Molecular Sciences, 19(1), 78. https://doi.org/10.3390/ijms19010078