Polyamine Metabolism and Gene Methylation in Conjunction with One-Carbon Metabolism
Abstract
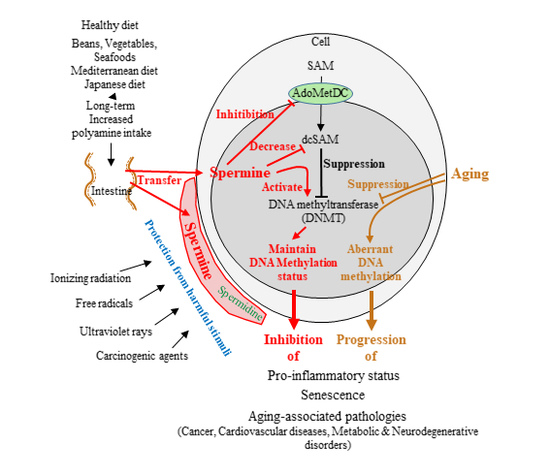
:1. Introduction
2. Aging-Associated Changes and Immunosenescence
3. Polyamines
4. Source of Body Polyamines
5. Dietary Polyamines
6. Epigenetics and Aging
7. Nutrients and Their Metabolites and Enzymes Related to DNA Methylation
7.1. Changing the Availability of Methyl Donors
7.2. Altering DNA Methyltransferase (DNMT) Activity
8. Aging, Polyamines, and DNA Methylation
9. Possible Role of Polyamines in Inhibiting Tumorigenesis
10. Future Perspectives
Funding
Conflicts of Interest
Abbreviations
| DNMT | DNA methyltransferase |
| SAM | S-adenosylmethionine |
| SAH | S-adenosyl-l-homocysteine |
| dcSAM | decarboxylated S-adenosylmethionine |
| CVD | cardiovascular disease |
| LFA-1 | lymphocyte function-associated antigen 1 |
| ODC | ornithine decarboxylase |
| AdoMetDC | adenosylmethionine decarboxylase |
| SSAT | spermidine/spermine N1-acetyltransferase |
| APAO | N1-acetylpolyamine oxidase |
| MTA | methylthioadenosine |
| DFMO | α-d,l-difluoromethylornithine hydrochloride |
References
- Soda, K.; Kano, Y.; Chiba, F. Food Polyamine and Cardiovascular Disease—An Epidemiological Study. Glob. J. Health Sci. 2012, 4, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Nagata, C.; Wada, K.; Tamura, T.; Konishi, K.; Goto, Y.; Koda, S.; Kawachi, T.; Tsuji, M.; Nakamura, K. Dietary Soy and Natto Intake and Cardiovascular Disease Mortality in Japanese Adults: The Takayama Study. Am. J. Clin Nutr. 2017, 105, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Erdman, J.W., Jr. Aha Science Advisory: Soy Protein and Cardiovascular Disease: A Statement for Healthcare Professionals from the Nutrition Committee of the Aha. Circulation 2000, 102, 2555–2559. [Google Scholar] [CrossRef] [PubMed]
- Trock, B.J.; Hilakivi-Clarke, L.; Clarke, R. Meta-Analysis of Soy Intake and Breast Cancer Risk. J. Natl. Cancer Inst. 2006, 98, 459–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, A.H.; Yu, M.C.; Tseng, C.C.; Pike, M.C. Epidemiology of Soy Exposures and Breast Cancer Risk. Br. J. Cancer 2008, 98, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Kim, J.H.; Nam, S.J.; Ryu, S.; Kong, G. Dietary Intake of Soy Protein and Tofu in Association with Breast Cancer Risk Based on a Case-Control Study. Nutr. Cancer 2008, 60, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.; Anthony, M.; Alexander, D.; Arab, L. Soy Consumption and Colorectal Cancer. Nutr. Cancer 2003, 47, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Oba, S.; Nagata, C.; Shimizu, N.; Shimizu, H.; Kametani, M.; Takeyama, N.; Ohnuma, T.; Matsushita, S. Soy Product Consumption and the Risk of Colon Cancer: A Prospective Study in Takayama, Japan. Nutr. Cancer 2007, 57, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Shu, X.O.; Li, H.; Chow, W.H.; Cai, H.; Zhang, X.; Gao, Y.T.; Zheng, W. Prospective Cohort Study of Soy Food Intake and Colorectal Cancer Risk in Women. Am. J. Clin. Nutr. 2009, 89, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Spitznagel, E.L.; Bosland, M.C. Soy Consumption and Colorectal Cancer Risk in Humans: A Meta-Analysis. Cancer Epidemiol. Biomarkers Prev. 2010, 19, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Sofi, F.; Abbate, R.; Gensini, G.F.; Casini, A. Accruing Evidence on Benefits of Adherence to the Mediterranean Diet on Health: An Updated Systematic Review and Meta-Analysis. Am. J. Clin. Nutr. 2010, 92, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Estruch, R.; Ros, E.; Salas-Salvadó, J.; Covas, M.I.; Corella, D.; Arós, F.; Gómez-Gracia, E.; Ruiz-Gutiérrez, V.; Fiol, M.; Lapetra, J.; et al. Primary Prevention of Cardiovascular Disease with a Mediterranean Diet Supplemented with Extra-Virgin Olive Oil or Nuts. N. Engl. J. Med. 2018, 378, e34. [Google Scholar] [CrossRef] [PubMed]
- Kris-Etherton, P.; Eckel, R.H.; Howard, B.V.; St Jeor, S.; Bazzarre, T.L. Nutrition Committee Population Science Committee; Clinical Science Committee of the American Heart Association. Aha Science Advisory: Lyon Diet Heart Study. Benefits of a Mediterranean-Style, National Cholesterol Education Program/American Heart Association Step I Dietary Pattern on Cardiovascular Disease. Circulation 2001, 103, 1823–1825. [Google Scholar] [PubMed]
- Benetou, V.; Trichopoulou, A.; Orfanos, P.; Naska, A.; Lagiou, P.; Boffetta, P.; Trichopoulos, D.; Greek, E.C. Conformity to Traditional Mediterranean Diet and Cancer Incidence: The Greek Epic Cohort. Br. J. Cancer 2008, 99, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Couto, E.; Boffetta, P.; Lagiou, P.; Ferrari, P.; Buckland, G.; Overvad, K.; Dahm, C.C.; Tjønneland, A.; Olsen, A.; Clavel-Chapelon, F.; et al. Mediterranean Dietary Pattern and Cancer Risk in the Epic Cohort. Br. J. Cancer 2011, 104, 1493–1499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trichopoulou, A.; Bamia, C.; Lagiou, P.; Trichopoulos, D. Conformity to Traditional Mediterranean Diet and Breast Cancer Risk in the Greek Epic (European Prospective Investigation into Cancer and Nutrition) Cohort. Am. J. Clin. Nutr. 2010, 92, 620–625. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.T.; Hu, F.B.; McCullough, M.L.; Newby, P.K.; Willett, W.C.; Holmes, M.D. Diet Quality Is Associated with the Risk of Estrogen Receptor-Negative Breast Cancer in Postmenopausal Women. J. Nutr. 2006, 136, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, M.; Rodriguez, E.; Birerdinc, A.; Baranova, A. Age-Independent Rise of Inflammatory Scores May Contribute to Accelerated Aging in Multi-Morbidity. Oncotarget 2015, 6, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Soda, K.; Kano, Y.; Nakamura, T.; Kasono, K.; Kawakami, M.; Konishi, F. Spermine, a Natural Polyamine, Suppresses LFA-1 Expression on Human Lymphocyte. J. Immunol. 2005, 175, 237–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrucci, L.; Corsi, A.; Lauretani, F.; Bandinelli, S.; Bartali, B.; Taub, D.D.; Guralnik, J.M.; Longo, D.L. The Origins of Age-Related Proinflammatory State. Blood 2005, 105, 2294–2299. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Campisi, J. Chronic Inflammation (Inflammaging) and Its Potential Contribution to Age-Associated Diseases. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69 (Suppl. 1), S4–S9. [Google Scholar] [CrossRef]
- Balmir, F.; Staack, R.; Jeffrey, E.; Jimenez, M.D.; Wang, L.; Potter, S.M. An extract of soy flour influences serum cholesterol and thyroid hormones in rats and hamsters. J. Nutr. 1996, 126, 3046–3053. [Google Scholar] [CrossRef] [PubMed]
- Sacks, F.M.; Lichtenstein, A.; Van Horn, L.; Harris, W.; Kris-Etherton, P.; Winston, M. Soy protein, isoflavones, and cardiovascular health: An american heart association science advisory for professionals from the nutrition committee. Circulation 2006, 113, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Lee, S.O.; Murphy, P.A.; Hendrich, S. Soy Protein with or without Isoflavones, soy germ and soy germ extract, and daidzein lessen plasma cholesterol levels in golden syrian hamsters. Exp. Biol. Med. 2003, 228, 1063–1068. [Google Scholar] [CrossRef]
- Baur, J.A.; Pearson, K.J.; Price, N.L.; Jamieson, H.A.; Lerin, C.; Kalra, A.; Prabhu, V.V.; Allard, J.S.; Lopez-Lluch, G.; Lewis, K.; et al. Resveratrol Improves Health and Survival of Mice on a High-Calorie Diet. Nature 2006, 444, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, R.; del Valle, J.; Modol, L.; Martinez, A.; Granado-Serrano, A.B.; Ramirez-Nunez, O.; Pallas, M.; Portero-Otin, M.; Osta, R.; Navarro, X. Resveratrol Improves Motoneuron Function and Extends Survival in Sod1(G93a) Als Mice. Neurotherapeutics 2014, 11, 419–432. [Google Scholar] [PubMed]
- Pearson, K.J.; Baur, J.A.; Lewis, K.N.; Peshkin, L.; Price, N.L.; Labinskyy, N.; Swindell, W.R.; Kamara, D.; Minor, R.K.; Perez, E.; et al. Resveratrol Delays Age-Related Deterioration and Mimics Transcriptional Aspects of Dietary Restriction without Extending Life Span. Cell Metab. 2008, 8, 157–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, R.A.; Harrison, D.E.; Astle, C.M.; Baur, J.A.; Boyd, A.R.; de Cabo, R.; Fernandez, E.; Flurkey, K.; Javors, M.A.; Nelson, J.F.; et al. Rapamycin, but Not Resveratrol or Simvastatin, Extends Life Span of Genetically Heterogeneous Mice. J. Gerontol. A Biol. Sci. Med. Sci. 2011, 66, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Staats, S.; Wagner, A.E.; Kowalewski, B.; Rieck, F.T.; Soukup, S.T.; Kulling, S.E.; Rimbach, G. Dietary Resveratrol Does Not Affect Life Span, Body Composition, Stress Response, and Longevity-Related Gene Expression in Drosophila Melanogaster. Int. J. Mol. Sci. 2018, 19, 223. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.A.; Riehle, M.A. Resveratrol Fails to Extend Life Span in the Mosquito Anopheles Stephensi. Rejuvenation Res. 2015, 18, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, M.; McDonagh, T.; Heltweg, B.; Hixon, J.; Westman, E.A.; Caldwell, S.D.; Napper, A.; Curtis, R.; DiStefano, P.S.; Fields, S.; et al. Substrate-Specific Activation of Sirtuins by Resveratrol. J. Biol. Chem. 2005, 280, 17038–17045. [Google Scholar] [CrossRef] [PubMed]
- Burnett, C.; Valentini, S.; Cabreiro, F.; Goss, M.; Somogyvári, M.; Piper, M.D.; Hoddinott, M.; Sutphin, G.L.; Leko, V.; McElwee, J.J.; et al. Absence of Effects of Sir2 Overexpression on Lifespan in C. Elegans and Drosophila. Nature 2011, 477, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Strong, R.; Miller, R.A.; Astle, C.M.; Baur, J.A.; de Cabo, R.; Fernandez, E.; Guo, W.; Javors, M.; Kirkland, J.L.; Nelson, J.F.; et al. Evaluation of Resveratrol, Green Tea Extract, Curcumin, Oxaloacetic Acid, and Medium-Chain Triglyceride Oil on Life Span of Genetically Heterogeneous Mice. J. Gerontol. A Biol. Sci. Med. Sci. 2013, 68, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Ernst, I.M.; Pallauf, K.; Bendall, J.K.; Paulsen, L.; Nikolai, S.; Huebbe, P.; Roeder, T.; Rimbach, G. Vitamin E Supplementation and Lifespan in Model Organisms. Ageing Res. Rev. 2013, 12, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.R.; Albert, C.M.; Gaziano, J.M.; Zaharris, E.; MacFadyen, J.; Danielson, E.; Buring, J.E.; Manson, J.E. A Randomized Factorial Trial of Vitamins C and E and β Carotene in the Secondary Prevention of Cardiovascular Events in Women: Results from the Women’s Antioxidant Cardiovascular Study. Arch. Intern. Med. 2007, 167, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Fujii, K.; Yao, J.; Kishida, H.; Hosoe, K.; Sawashita, J.; Takeda, T.; Mori, M.; Higuchi, K. Reduced Coenzyme Q10 Supplementation Decelerates Senescence in Samp1 Mice. Exp. Gerontol 2006, 41, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Pugh, T.D.; Klopp, R.G.; Edwards, J.; Allison, D.B.; Weindruch, R.; Prolla, T.A. The Impact of α-Lipoic Acid, Coenzyme Q10 and Caloric Restriction on Life Span and Gene Expression Patterns in Mice. Free Radic. Biol. Med. 2004, 36, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Miller, E.R., 3rd.; Pastor-Barriuso, R.; Dalal, D.; Riemersma, R.A.; Appel, L.J.; Guallar, E. Meta-Analysis: High-Dosage Vitamin E Supplementation May Increase All-Cause Mortality. Ann. Intern. Med. 2005, 142, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Vivekananthan, D.P.; Penn, M.S.; Sapp, S.K.; Hsu, A.; Topol, E.J. Use of Antioxidant Vitamins for the Prevention of Cardiovascular Disease: Meta-Analysis of Randomised Trials. Lancet 2003, 361, 2017–2023. [Google Scholar] [CrossRef]
- Hsieh, C.C.; Lin, B.F. Opposite Effects of Low and High Dose Supplementation of Vitamin E on Survival of Mrl/Lpr Mice. Nutrition 2005, 21, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Selman, C.; McLaren, J.S.; Meyer, C.; Duncan, J.S.; Redman, P.; Collins, A.R.; Duthie, G.G.; Speakman, J.R. Life-Long Vitamin C Supplementation in Combination with Cold Exposure Does Not Affect Oxidative Damage or Lifespan in Mice, but Decreases Expression of Antioxidant Protection Genes. Mech. Ageing Dev. 2006, 127, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Morley, A.A.; Trainor, K.J. Lack of an Effect of Vitamin E on Lifespan of Mice. Biogerontology 2001, 2, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.Q.; Yi, K.H.; Li, Z.; Wang, H.; Li, M.L.; Cai, L.L.; Lin, H.N.; Lin, Q.; Tzeng, C.M. DNA Methylation Profiling Reveals the Change of Inflammation-Associated Zc3h12d in Leukoaraiosis. Front. Aging Neurosci. 2018, 10, 143. [Google Scholar] [CrossRef] [PubMed]
- Irvin, M.R.; Aslibekyan, S.; Do, A.; Zhi, D.; Hidalgo, B.; Claas, S.A.; Srinivasasainagendra, V.; Horvath, S.; Tiwari, H.K.; Absher, D.M.; et al. Metabolic and Inflammatory Biomarkers Are Associated with Epigenetic Aging Acceleration Estimates in the Goldn Study. Clin. Epigenet. 2018, 10, 56. [Google Scholar] [CrossRef] [PubMed]
- Kano, Y.; Soda, K.; Konishi, F. Suppression of LFA-1 Expression by Spermine Is Associated with Enhanced Methylation of ITGAL, the LFA-1 Promoter Area. PLoS ONE 2013, 8, e56056. [Google Scholar] [CrossRef] [PubMed]
- Soda, K.; Dobashi, Y.; Kano, Y.; Tsujinaka, S.; Konishi, F. Polyamine-Rich Food Decreases Age-Associated Pathology and Mortality in Aged Mice. Exp. Gerontol. 2009, 44, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Soda, K.; Kano, Y.; Chiba, F.; Koizumi, K.; Miyaki, Y. Increased Polyamine Intake Inhibits Age-Associated Alteration in Global DNA Methylation and 1,2-Dimethylhydrazine-Induced Tumorigenesis. PLoS ONE 2013, 8, e64357. [Google Scholar] [CrossRef] [PubMed]
- Powers, D.C.; Morley, J.E.; Flood, J.F. Age-Related Changes in LFA-1 Expression, Cell Adhesion, and Pha-Induced Proliferation by Lymphocytes from Senescence-Accelerated Mouse (Sam)-P/8 and Sam-R/1 Substrains. Cell Immunol. 1992, 141, 444–456. [Google Scholar] [CrossRef]
- Pallis, M.; Robins, A.; Powell, R. Quantitative Analysis of Lymphocyte Cd11a Using Standardized Flow Cytometry. Scand. J. Immunol. 1993, 38, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Okumura, M.; Fujii, Y.; Takeuchi, Y.; Inada, K.; Nakahara, K.; Matsuda, H. Age-Related Accumulation of LFA-1high Cells in a CD8+ CD45RAhigh T Cell Population. Eur. J. Immunol. 1993, 23, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Chiricolo, M.; Morini, M.C.; Mancini, R.; Beltrandi, E.; Belletti, D.; Conte, R. Cell Adhesion Molecules Cd11a and Cd18 in Blood Monocytes in Old Age and the Consequences for Immunological Dysfunction. Preliminary Results. Gerontology 1995, 41, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Arias, R.; Moro-Garcia, M.A.; Lopez-Vazquez, A.; Rodrigo, L.; Baltar, J.; Garcia, F.M.; Jaurrieta, J.J.; Lopez-Larrea, C. Nkg2d Expression in CD4+ T Lymphocytes as a Marker of Senescence in the Aged Immune System. Age 2011, 33, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Sandmand, M.; Bruunsgaard, H.; Kemp, K.; Andersen-Ranberg, K.; Pedersen, A.N.; Skinhoj, P.; Pedersen, B.K. Is Ageing Associated with a Shift in the Balance between Type 1 and Type 2 Cytokines in Humans? Clin. Exp. Immunol. 2002, 127, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Li, G.; Lee, W.W.; Yuan, M.; Cui, D.; Weyand, C.M.; Goronzy, J.J. Signal Inhibition by the Dual-Specific Phosphatase 4 Impairs T Cell-Dependent B-Cell Responses with Age. Proc. Natl Acad. Sci. USA 2012, 109, E879–E888. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Bonafe, M.; Valensin, S.; Olivieri, F.; De Luca, M.; Ottaviani, E.; De Benedictis, G. Inflamm-Aging. An Evolutionary Perspective on Immunosenescence. Ann. N. Y. Acad. Sci. 2000, 908, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Flesher, D.L.; Sun, X.; Behrens, T.W.; Graham, R.R.; Criswell, L.A. Recent Advances in the Genetics of Systemic Lupus Erythematosus. Expert Rev. Clin. Immunol. 2010, 6, 461–479. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Sridharan, A.; Prakash, S.; Agrawal, H. Dendritic Cells and Aging: Consequences for Autoimmunity. Expert Rev. Clin. Immunol. 2012, 8, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Sung, B.; Aggarwal, B.B. Age-Associated Chronic Diseases Require Age-Old Medicine: Role of Chronic Inflammation. Prev. Med. 2012, 54, S29–S37. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.C.; McCusker, R.H.; Strle, K.; Johnson, R.W.; Dantzer, R.; Kelley, K.W. Regulation of IGF-I Function by Proinflammatory Cytokines: At the Interface of Immunology and Endocrinology. Cell Immunol. 2008, 252, 91–110. [Google Scholar] [CrossRef] [PubMed]
- De Luca, C.; Olefsky, J.M. Inflammation and Insulin Resistance. FEBS Lett. 2008, 582, 97–105. [Google Scholar] [CrossRef] [PubMed]
- La Ferla, K.; Reimann, C.; Jelkmann, W.; Hellwig-Burgel, T. Inhibition of Erythropoietin Gene Expression Signaling Involves the Transcription Factors Gata-2 and NF-κB. FASEB J. 2002, 16, 1811–1813. [Google Scholar] [CrossRef] [PubMed]
- Sprague, A.H.; Khalil, R.A. Inflammatory Cytokines in Vascular Dysfunction and Vascular Disease. Biochem. Pharmacol. 2009, 78, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Canoves, P.; Scheele, C.; Pedersen, B.K.; Serrano, A.L. Interleukin-6 Myokine Signaling in Skeletal Muscle: A Double-Edged Sword? FEBS J. 2013, 280, 4131–4148. [Google Scholar] [CrossRef] [PubMed]
- Fulop, T.; Larbi, A.; Pawelec, G. Human T Cell Aging and the Impact of Persistent Viral Infections. Front. Immunol. 2013, 4, 271. [Google Scholar] [CrossRef] [PubMed]
- Pawelec, G. Immunosenenescence: Role of Cytomegalovirus. Exp. Gerontol. 2014, 54, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ferioli, M.E.; Ceruti, G.; Comolli, R. Changes in Rat Liver Ornithine Decarboxylase Activity During Ageing and Effect of Stimulation by Dexamethasone. Exp. Gerontol. 1976, 11, 153–156. [Google Scholar] [CrossRef]
- Yoshinaga, K.; Ishizuka, J.; Evers, B.M.; Townsend, C.M., Jr.; Thompson, J.C. Age-Related Changes in Polyamine Biosynthesis after Fasting and Refeeding. Exp. Gerontol. 1993, 28, 565–572. [Google Scholar] [CrossRef]
- Janne, J.; Raina, A. On the Stimulation of Ornithine Decarboxylase and Rna Polymerase Activity in Rat Liver after Treatment with Growth Hormone. Biochim. Biophys. Acta. 1969, 174, 769–772. [Google Scholar] [CrossRef]
- Bedford, M.R.; Smith, T.K.; Summers, J.D. Effect of Dietary Ornithine on Renal and Hepatic Polyamine Synthesis. Ann. Nutr. Metab. 1988, 32, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Schleiffer, R.; Duranton, B.; Gosse, F.; Hasselmann, M.; Raul, F. Blood Polyamine Levels after Oral Ornithine Load, a Diagnostic Marker of Hyperproliferative Premalignant and Malignant Stages in a Model of Colon Carcinogenesis. Cancer Detect. Prev. 2000, 24, 542–548. [Google Scholar] [PubMed]
- Teixeira, D.; Santaolaria, M.L.; Meneu, V.; Alonso, E. Dietary Arginine Slightly and Variably Affects Tissue Polyamine Levels in Male Swiss Albino Mice. J. Nutr. 2002, 132, 3715–3720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laitinen, S.I.; Laitinen, P.H.; Hietala, O.A.; Pajunen, A.E.; Piha, R.S. Developmental Changes in Mouse Brain Polyamine Metabolism. Neurochem. Res. 1982, 7, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Kanungo, M.S. Activity and Modulation of Ornithine Decarboxylase and Concentrations of Polyamines in Various Tissues of Rats as a Function of Age. Exp. Gerontol. 1982, 17, 95–103. [Google Scholar] [CrossRef]
- Zhang, M.; Caragine, T.; Wang, H.; Cohen, P.S.; Botchkina, G.; Soda, K.; Bianchi, M.; Ulrich, P.; Cerami, A.; Sherry, B.; et al. Spermine Inhibits Proinflammatory Cytokine Synthesis in Human Mononuclear Cells: A Counterregulatory Mechanism That Restrains the Immune Response. J. Exp. Med. 1997, 185, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Gillis, S.; Kozak, R.; Durante, M.; Weksler, M.E. Immunological Studies of Aging. Decreased Production of and Response to T Cell Growth Factor by Lymphocytes from Aged Humans. J. Clin. Investig. 1981, 67, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Soda, K. Anti-Aging by Polyamine. Food Style 21 2006, 10, 43–54. Available online: http://www.natto.or.jp/thesis/01polyamine/07.html (accessed on 9 Oct 2018). (In Japanese).
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of Autophagy by Spermidine Promotes Longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Soda, K. The Mechanisms by Which Polyamines Accelerate Tumor Spread. J. Exp. Clin. Cancer Res. 2011, 30, 95. [Google Scholar] [CrossRef] [PubMed]
- Weiss, T.S.; Bernhardt, G.; Buschauer, A.; Thasler, W.E.; Dolgner, D.; Zirngibl, H.; Jauch, K.W. Polyamine levels of human colorectal adenocarcinomas are correlated with tumor stage and grade. Int. J. Colorectal Dis. 2002, 17, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Upp, J.R., Jr.; Saydjari, R.; Townsend, C.M., Jr.; Singh, P.; Barranco, S.C.; Thompson, J.C. Polyamine Levels and Gastrin Receptors in Colon Cancers. Ann. Surg. 1988, 207, 662–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallesio, C.; Colombatto, S.; Modica, R. Free and Acetylated Polyamines as Markers of Oral Cavity Tumors. Oral Surg. Oral Med. Oral Pathol. 1994, 77, 167–171. [Google Scholar] [CrossRef]
- Becciolini, A.; Porciani, S.; Lanini, A.; Balzi, M.; Cionini, L.; Bandettini, L. Polyamine Levels in Healthy and Tumor Tissues of Patients with Colon Adenocarcinoma. Dis. Colon Rectum 1991, 34, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Loser, C.; Folsch, U.R.; Paprotny, C.; Creutzfeldt, W. Polyamines in Colorectal Cancer. Evaluation of Polyamine Concentrations in the Colon Tissue, Serum, and Urine of 50 Patients with Colorectal Cancer. Cancer 1990, 65, 958–966. [Google Scholar] [CrossRef]
- Elworthy, P.; Hitchcock, E. Polyamine Levels in Red Blood Cells from Patient Groups of Different Sex and Age. Biochim. Biophys. Acta 1989, 993, 212–216. [Google Scholar] [CrossRef]
- Cipolla, B.G.; Havouis, R.; Moulinoux, J.P. Polyamine Contents in Current Foods: A Basis for Polyamine Reduced Diet and a Study of Its Long Term Observance and Tolerance in Prostate Carcinoma Patients. Amino Acids 2007, 33, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Nishibori, N.; Fujihara, S.; Akatuki, T. Amounts of Polyamines in Foods in Japan and Intake by Japanese. Food Chem. 2006, 100. [Google Scholar] [CrossRef]
- Nishimura, K.; Shiina, R.; Kashiwagi, K.; Igarashi, K. Decrease in Polyamines with Aging and Their Ingestion from Food and Drink. J. Biochem. 2006, 139, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Soda, K.; Mogi, S.; Shiina, M.; Kawabata, N. The Polyamine Content in Various Foods on a Calorie Basis. JACOBS J. Food Nutr. 2017, 4, 029. [Google Scholar]
- Uda, K.; Tsujikawa, T.; Fujiyama, Y.; Bamba, T. Rapid Absorption of Luminal Polyamines in a Rat Small Intestine Ex Vivo Model. J. Gastroenterol. Hepatol. 2003, 18, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Bardocz, S.; Brown, D.S.; Grant, G.; Pusztai, A. Luminal and Basolateral Polyamine Uptake by Rat Small Intestine Stimulated to Grow by Phaseolus Vulgaris Lectin Phytohaemagglutinin In Vivo. Biochim. Biophys. Acta 1990, 1034, 46–52. [Google Scholar] [CrossRef]
- Bardocz, S.; Duguid, T.J.; Brown, D.S.; Grant, G.; Pusztai, A.; White, A.; Ralph, A. The Importance of Dietary Polyamines in Cell Regeneration and Growth. Br. J. Nutr. 1995, 73, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Cipolla, B.; Guilli, F.; Moulinoux, J.P. Polyamine-Reduced Diet in Metastatic Hormone-Refractory Prostate Cancer (Hrpc) Patients. Biochem. Soc. Trans. 2003, 31, 384–387. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Araki, N.; Ohnishi, Y.; Kozaki, S. Effects of Dietary Polyamine Deficiency on Trypanosoma Gambiense Infection in Rats. Exp. Parasitol. 2001, 97, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Sarhan, S.; Knodgen, B.; Seiler, N. The Gastrointestinal Tract as Polyamine Source for Tumor Growth. Anticancer Res. 1989, 9, 215–223. [Google Scholar] [PubMed]
- Soda, K.; Kano, Y.; Sakuragi, M.; Takao, K.; Lefor, A.; Konishi, F. Long-Term Oral Polyamine Intake Increases Blood Polyamine Concentrations. J. Nutr. Sci. Vitaminol. 2009, 55, 361–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodal, B.P.; Eliassen, K.A.; Ronning, H.; Osmundsen, H. Effects of Dietary Polyamines and Clofibrate on Metabolism of Polyamines in the Rat. J. Nutr. Biochem. 1999, 10, 700–708. [Google Scholar] [CrossRef]
- Soda, K.; Uemura, T.; Igarashi, K.; Fukui, T. Increased Polyamine Intake by Being Adherent to Traditional Japanese Diet Increases Blood Spermine Levels and Inhibits Pro-Inflammatory Status—An Interventional Study. Unpublished.
- Yuan, Q.; Ray, R.M.; Viar, M.J.; Johnson, L.R. Polyamine Regulation of Ornithine Decarboxylase and Its Antizyme in Intestinal Epithelial Cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 280, G130–G138. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Abdellatif, M.; Schroeder, S.; Primessnig, U.; Stekovic, S.; Pendl, T.; Harger, A.; Schipke, J.; Zimmermann, A.; Schmidt, A.; et al. Cardioprotection and Lifespan Extension by the Natural Polyamine Spermidine. Nat. Med. 2016, 22, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Jiang, J.; Yu, P.; Zhang, G.; Zhang, G.; Liu, X. Green Tea Polyphenol Treatment Attenuates Atherosclerosis in High-Fat Diet-Fed Apolipoprotein E-Knockout Mice Via Alleviating Dyslipidemia and up-Regulating Autophagy. PLoS ONE 2017, 12, e0181666. [Google Scholar] [CrossRef] [PubMed]
- Ferraresi, A.; Phadngam, S.; Morani, F.; Galetto, A.; Alabiso, O.; Chiorino, G.; Isidoro, C. Resveratrol Inhibits Il-6-Induced Ovarian Cancer Cell Migration through Epigenetic up-Regulation of Autophagy. Mol. Carcinog. 2017, 56, 1164–1181. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Jeong, H.; Lee, M.N.; Koh, A.; Kwon, O.; Yang, Y.R.; Noh, J.; Suh, P.G.; Park, H.; Ryu, S.H. Resveratrol Induces Autophagy by Directly Inhibiting mTOR through ATP Competition. Sci. Rep. 2016, 6, 21772. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Yang, F.; Fang, Z.; Hu, C. Resveratrol Ameliorates Alcoholic Fatty Liver by Inducing Autophagy. Am. J. Chin. Med. 2016, 44, 1207–1220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Xu, J.; Lu, Y.; Jiang, J.; Wang, L.; Shen, H.M.; Xia, D. Curcumin Targets the Tfeb-Lysosome Pathway for Induction of Autophagy. Oncotarget 2016, 7, 75659–75671. [Google Scholar] [CrossRef] [PubMed]
- Pucciarelli, S.; Moreschini, B.; Micozzi, D.; De Fronzo, G.S.; Carpi, F.M.; Polzonetti, V.; Vincenzetti, S.; Mignini, F.; Napolioni, V. Spermidine and Spermine Are Enriched in Whole Blood of Nona/Centenarians. Rejuvenation Res. 2012, 15, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Sato, S.; Nagase, S.; Shimosato, K.; Ohkuma, S. Effects of Methotrexate and Cyclophosphamide on Polyamine Levels in Various Tissues of Rats. J. Drug Target 1999, 7, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Binh, P.N.T.; Soda, K.; Kawakami, M. Mediterranean Diet and Polyamine Intake: Possible Contribution of Increased Polyamine Intake to Inhibition of Age-Associated Disease. Nutr. Diet. Suppl. 2011, 3, 1–7. [Google Scholar]
- Fukui, T.; Soda, K.; Takao, K.; Rikiyama, T. Extracellular spermine activates DNA methyltransferase 3A and 3B. Unpublished.
- Bestor, T.; Laudano, A.; Mattaliano, R.; Ingram, V. Cloning and Sequencing of a Cdna Encoding DNA Methyltransferase of Mouse Cells. The Carboxyl-Terminal Domain of the Mammalian Enzymes Is Related to Bacterial Restriction Methyltransferases. J. Mol. Biol. 1988, 203, 971–983. [Google Scholar] [CrossRef]
- Garcea, R.; Daino, L.; Pascale, R.; Simile, M.M.; Puddu, M.; Ruggiu, M.E.; Seddaiu, M.A.; Satta, G.; Sequenza, M.J.; Feo, F. Protooncogene Methylation and Expression in Regenerating Liver and Preneoplastic Liver Nodules Induced in the Rat by Diethylnitrosamine: Effect of Variations of S-Adenosylmethionine: S-Adenosylhomocysteine Ratio. Carcinogenesis 1989, 10, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Lovaas, E.; Carlin, G. Spermine: An Anti-Oxidant and Anti-Inflammatory Agent. Free Radic. Biol. Med. 1991, 11, 455–461. [Google Scholar] [CrossRef]
- Lagishetty, C.V.; Naik, S.R. Polyamines: Potential Anti-Inflammatory Agents and Their Possible Mechanism of Action. Indian J. Pharmacol. 2008, 40, 121–125. [Google Scholar] [PubMed]
- Choi, Y.H.; Park, H.Y. Anti-Inflammatory Effects of Spermidine in Lipopolysaccharide-Stimulated Bv2 Microglial Cells. J. Biomed. Sci. 2012, 19, 31. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Kang, S.C. Natural Polyamine Inhibits Mouse Skin Inflammation and Macrophage Activation. Inflamm. Res. 2013, 62, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Gu, J.; Liu, R.; Wei, S.; Wang, Q.; Shen, H.; Dai, Y.; Zhou, H.; Zhang, F.; Lu, L. Spermine Alleviates Acute Liver Injury by Inhibiting Liver-Resident Macrophage Pro-Inflammatory Response through Atg5-Dependent Autophagy. Front. Immunol. 2018, 9, 948. [Google Scholar] [CrossRef] [PubMed]
- Tadolini, B.; Cabrini, L.; Landi, L.; Varani, E.; Pasquali, P. Polyamine Binding to Phospholipid Vesicles and Inhibition of Lipid Peroxidation. Biochem. Biophys. Res. Commun. 1984, 122, 550–555. [Google Scholar] [CrossRef]
- Khan, A.U.; Di Mascio, P.; Medeiros, M.H.; Wilson, T. Spermine and Spermidine Protection of Plasmid DNA against Single-Strand Breaks Induced by Singlet Oxygen. Proc. Natl. Acad. Sci. USA 1992, 89, 11428–11430. [Google Scholar] [CrossRef] [PubMed]
- Goss, S.P.; Hogg, N.; Kalyanaraman, B. The Antioxidant Effect of Spermine Nonoate in Human Low-Density Lipoprotein. Chem. Res. Toxicol. 1995, 8, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Marzabadi, M.R.; Llvaas, E. Spermine Prevent Iron Accumulation and Depress Lipofuscin Accumulation in Cultured Myocardial Cells. Free Radic. Biol. Med. 1996, 21, 375–381. [Google Scholar] [CrossRef]
- Farbiszewski, R.; Bielawska, A.; Szymanska, M.; Skrzydlewska, E. Spermine partially normalizes in vivo antioxidant defense potential in certain brain regions in transiently hypoperfused rat brain. Neurochem. Res. 1996, 21, 1497–1503. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.C.; Sirisoma, N.S.; Kuppusamy, P.; Zweier, J.L.; Woster, P.M.; Casero, R.A., Jr. The Natural Polyamine Spermine Functions Directly as a Free Radical Scavenger. Proc. Natl. Acad. Sci. USA 1998, 95, 11140–11145. [Google Scholar] [CrossRef] [PubMed]
- Jung, I.L.; Oh, T.J.; Kim, I.G. Abnormal Growth of Polyamine-Deficient Escherichia Coli Mutant Is Partially Caused by Oxidative Stress-Induced Damage. Arch. Biochem. Biophys. 2003, 418, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, M.K.; Tabor, C.W.; Tabor, H. Polyamines Protect Escherichia Coli Cells from the Toxic Effect of Oxygen. Proc. Natl. Acad. Sci. USA 2003, 100, 2261–2265. [Google Scholar] [CrossRef] [PubMed]
- Belle, N.A.; Dalmolin, G.D.; Fonini, G.; Rubin, M.A.; Rocha, J.B. Polyamines Reduces Lipid Peroxidation Induced by Different Pro-Oxidant Agents. Brain Res. 2004, 1008, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Gaboriau, F.; Vaultier, M.; Moulinoux, J.P.; Delcros, J.G. Antioxidative Properties of Natural Polyamines and Dimethylsilane Analogues. Redox. Rep. 2005, 10, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, S.; Kadoma, Y. Kinetic Evaluation of Polyamines as Radical Scavengers. Anticancer Res. 2005, 25, 965–969. [Google Scholar] [PubMed]
- Sava, I.G.; Battaglia, V.; Rossi, C.A.; Salvi, M.; Toninello, A. Free Radical Scavenging Action of the Natural Polyamine Spermine in Rat Liver Mitochondria. Free Radic. Biol. Med. 2006, 41, 1272–1281. [Google Scholar] [CrossRef] [PubMed]
- Rider, J.E.; Hacker, A.; Mackintosh, C.A.; Pegg, A.E.; Woster, P.M.; Casero, R.A., Jr. Spermine and Spermidine Mediate Protection against Oxidative Damage Caused by Hydrogen Peroxide. Amino Acids 2007, 33, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Nayvelt, I.; Hyvonen, M.T.; Alhonen, L.; Pandya, I.; Thomas, T.; Khomutov, A.R.; Vepsalainen, J.; Patel, R.; Keinanen, T.A.; Thomas, T.J. DNA Condensation by Chiral Alpha-Methylated Polyamine Analogues and Protection of Cellular DNA from Oxidative Damage. Biomacromolecules 2010, 11, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.W.; Cha, H.J.; Han, M.H.; Hwang, S.J.; Lee, D.S.; Yoo, J.S.; Choi, I.W.; Kim, S.; Kim, H.S.; Kim, G.Y.; et al. Spermidine Protects against Oxidative Stress in Inflammation Models Using Macrophages and Zebrafish. Biomol. Ther. 2018, 26, 146–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courdi, A.; Milano, G.; Bouclier, M.; Lalanne, C.M. Radiosensitization of Human Tumor Cells by Alpha-Difluoromethylornithine. Int. J. Cancer 1986, 38, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Arundel, C.M.; Nishioka, K.; Tofilon, P.J. Effects of Alpha-Difluoromethylornithine-Induced Polyamine Depletion on the Radiosensitivity of a Human Colon Carcinoma Cell Line. Radiat. Res. 1988, 114, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Held, K.D.; Awad, S. Effects of Polyamines and Thiols on the Radiation Sensitivity of Bacterial Transforming DNA. Int. J. Radiat. Biol. 1991, 59, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Snyder, R.D.; Schroeder, K.K. Radiosensitivity of Polyamine-Depleted Hela Cells and Modulation by the Aminothiol Wr-1065. Radiat. Res. 1994, 137, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.R.; Casero, R.A.; Dillehay, L.E. The Effect of Polyamine Depletion on the Cytotoxic Response to Puva, Gamma Rays and Uvc in V79 Cells in Vitro. Biochem. Biophys. Res. Commun. 1994, 201, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Spotheim-Maurizot, M.; Ruiz, S.; Sabattier, R.; Charlier, M. Radioprotection of DNA by Polyamines. Int. J. Radiat. Biol. 1995, 68, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Newton, G.L.; Aguilera, J.A.; Ward, J.F.; Fahey, R.C. Polyamine-Induced Compaction and Aggregation of DNA—A Major Factor in Radioprotection of Chromatin under Physiological Conditions. Radiat. Res. 1996, 145, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Chiu, S.; Oleinick, N.L. Radioprotection of Cellular Chromatin by the Polyamines Spermine and Putrescine: Preferential Action against Formation of DNA-Protein Crosslinks. Radiat. Res. 1998, 149, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Sy, D.; Hugot, S.; Savoye, C.; Ruiz, S.; Charlier, M.; Spotheim-Maurizot, M. Radioprotection of DNA by Spermine: A Molecular Modelling Approach. Int. J. Radiat. Biol. 1999, 75, 953–961. [Google Scholar] [PubMed]
- Warters, R.L.; Newton, G.L.; Olive, P.L.; Fahey, R.C. Radioprotection of Human Cell Nuclear DNA by Polyamines: Radiosensitivity of Chromatin Is Influenced by Tightly Bound Spermine. Radiat. Res. 1999, 151, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Douki, T.; Bretonniere, Y.; Cadet, J. Protection against Radiation-Induced Degradation of DNA Bases by Polyamines. Radiat. Res. 2000, 153, 29–35. [Google Scholar] [CrossRef]
- Von Deutsch, A.W.; Mitchell, C.D.; Williams, C.E.; Dutt, K.; Silvestrov, N.A.; Klement, B.J.; Abukhalaf, I.K.; von Deutsch, D.A. Polyamines Protect against Radiation-Induced Oxidative Stress. Gravit. Space Biol. Bull. 2005, 18, 109–110. [Google Scholar] [PubMed]
- Snyder, R.D.; Sunkara, P.S. Effect of Polyamine Depletion on DNA Damage and Repair Following Uv Irradiation of Hela Cells. Photochem. Photobiol. 1990, 52, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Rajalakshmi, S.; Rao, P.M.; Sarma, D.S. Studies on Carcinogen Chromatin—DNA Interaction: Inhibition of N-Methyl-N-Nitrosourea-Induced Methylation of Chromatin—DNA by Spermine and Distamycin A. Biochemistry 1978, 17, 4515–4518. [Google Scholar] [CrossRef] [PubMed]
- Pothipongsa, A.; Jantaro, S.; Incharoensakdi, A. Polyamines Induced by Osmotic Stress Protect Synechocystis Sp. Pcc 6803 Cells and Arginine Decarboxylase Transcripts against Uv-B Radiation. Appl. Biochem. Biotechnol. 2012, 168, 1476–1488. [Google Scholar] [CrossRef] [PubMed]
- Mackintosh, C.A.; Pegg, A.E. Effect of Spermine Synthase Deficiency on Polyamine Biosynthesis and Content in Mice and Embryonic Fibroblasts, and the Sensitivity of Fibroblasts to 1,3-Bis-(2-Chloroethyl)-N-Nitrosourea. Biochem. J. 2000, 351, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Di Mascio, P.; Teixeira, P.C.; Onuki, J.; Medeiros, M.H.; Dornemann, D.; Douki, T.; Cadet, J. DNA Damage by 5-Aminolevulinic and 4,5-Dioxovaleric Acids in the Presence of Ferritin. Arch. Biochem. Biophys. 2000, 373, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, S.D.; Seggara, G.; Vo, P.A.; Macallister, R.J.; Hobbs, A.J.; Ahluwalia, A. Protection against Lipopolysaccharide-Induced Endothelial Dysfunction in Resistance and Conduit Vasculature of Inos Knockout Mice. FASEB J. 2003, 17, 773–775. [Google Scholar] [CrossRef] [PubMed]
- Gugliucci, A.; Menini, T. The Polyamines Spermine and Spermidine Protect Proteins from Structural and Functional Damage by Age Precursors: A New Role for Old Molecules? Life Sci. 2003, 72, 2603–2616. [Google Scholar] [CrossRef]
- Sagor, G.H.; Berberich, T.; Takahashi, Y.; Niitsu, M.; Kusano, T. The Polyamine Spermine Protects Arabidopsis from Heat Stress-Induced Damage by Increasing Expression of Heat Shock-Related Genes. Transgenic Res. 2013, 22, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Okumura, S.; Teratani, T.; Fujimoto, Y.; Zhao, X.; Tsuruyama, T.; Masano, Y.; Kasahara, N.; Iida, T.; Yagi, S.; Uemura, T.; et al. Oral Administration of Polyamines Ameliorates Liver Ischemia/Reperfusion Injury and Promotes Liver Regeneration in Rats. Liver Transpl. 2016, 22, 1231–1244. [Google Scholar] [CrossRef] [PubMed]
- Kucharski, R.; Maleszka, J.; Foret, S.; Maleszka, R. Nutritional Control of Reproductive Status in Honeybees Via DNA Methylation. Science 2008, 319, 1827–1830. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shangguan, S.; Xin, Y.; Chang, S.; Wang, Z.; Lu, X.; Wu, L.; Niu, B.; Zhang, T. Folate Deficiency Disturbs Hsa-Let-7 G Level through Methylation Regulation in Neural Tube Defects. J. Cell Mol. Med. 2017, 21, 3244–3253. [Google Scholar] [CrossRef] [PubMed]
- Toriyama, M.; Toriyama, M.; Wallingford, J.B.; Finnell, R.H. Folate-Dependent Methylation of Septins Governs Ciliogenesis During Neural Tube Closure. FASEB J. 2017, 31, 3622–3635. [Google Scholar] [CrossRef] [PubMed]
- Degroote, S.; Hunting, D.; Takser, L. Periconceptional Folate Deficiency Leads to Autism-Like Traits in Wistar Rat Offspring. Neurotoxicol. Teratol. 2018, 66, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Fekete, K.; Berti, C.; Cetin, I.; Hermoso, M.; Koletzko, B.V.; Decsi, T. Perinatal Folate Supply: Relevance in Health Outcome Parameters. Matern. Child. Nutr. 2010, 6 (Suppl. 2), 23–38. [Google Scholar] [CrossRef]
- Li, W.; Li, Z.; Li, S.; Wang, X.; Wilson, J.X.; Huang, G. Periconceptional Folic Acid Supplementation Benefit to Development of Early Sensory-Motor Function through Increase DNA Methylation in Rat Offspring. Nutrients 2018, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Zhuo, Y.; Wang, J.; Zhao, Y.; Xuan, Y.; Mou, D.; Liu, H.; Zhou, P.; Fang, Z.; Che, L.; et al. Methyl Donors Dietary Supplementation to Gestating Sows Diet Improves the Growth Rate of Offspring and Is Associating with Changes in Expression and DNA Methylation of Insulin-Like Growth Factor-1 Gene. J. Anim. Physiol. Anim. Nutr. 2018, 102, 1340–1350. [Google Scholar] [CrossRef] [PubMed]
- Steegers-Theunissen, R.P.; Obermann-Borst, S.A.; Kremer, D.; Lindemans, J.; Siebel, C.; Steegers, E.A.; Slagboom, P.E.; Heijmans, B.T. Periconceptional Maternal Folic Acid Use of 400 Microg Per Day Is Related to Increased Methylation of the IGF2 Gene in the Very Young Child. PLoS ONE 2009, 4, e7845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, W.N.; Khulan, B.; Owens, S.; Elks, C.E.; Seidel, V.; Prentice, A.M.; Belteki, G.; Ong, K.K.; Affara, N.A.; Constância, M.; et al. DNA Methylation Profiling at Imprinted Loci after Periconceptional Micronutrient Supplementation in Humans: Results of a Pilot Randomized Controlled Trial. FASEB J. 2012, 26, 1782–1790. [Google Scholar] [CrossRef] [PubMed]
- Joubert, B.R.; den Dekker, H.T.; Felix, J.F.; Bohlin, J.; Ligthart, S.; Beckett, E.; Tiemeier, H.; van Meurs, J.B.; Uitterlinden, A.G.; Hofman, A.; et al. Maternal Plasma Folate Impacts Differential DNA Methylation in an Epigenome-Wide Meta-Analysis of Newborns. Nat. Commun. 2016, 7, 10577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauwels, S.; Ghosh, M.; Duca, R.C.; Bekaert, B.; Freson, K.; Huybrechts, I.; Langie, S.A.S.; Koppen, G.; Devlieger, R.; Godderis, L. Dietary and Supplemental Maternal Methyl-Group Donor Intake and Cord Blood DNA Methylation. Epigenetics 2017, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, S.; Ghosh, M.; Duca, R.C.; Bekaert, B.; Freson, K.; Huybrechts, I.; Langie, S.A.S.; Koppen, G.; Devlieger, R.; Godderis, L. Maternal Intake of Methyl-Group Donors Affects DNA Methylation of Metabolic Genes in Infants. Clin. Epigenet. 2017, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Kochmanski, J.; Marchlewicz, E.H.; Cavalcante, R.G.; Sartor, M.A.; Dolinoy, D.C. Age-Related Epigenome-Wide DNA Methylation and Hydroxymethylation in Longitudinal Mouse Blood. Epigenetics 2018, 13, 779–792. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Deng, C.; Lu, Q.; Richardson, B. Age-Dependent DNA Methylation Changes in the ITGAL (Cd11a) Promoter. Mech. Ageing Dev. 2002, 123, 1257–1268. [Google Scholar] [CrossRef]
- Nguyen, A.; Leblond, F.; Mamarbachi, M.; Geoffroy, S.; Thorin, E. Age-Dependent Demethylation of Sod2 Promoter in the Mouse Femoral Artery. Oxid. Med. Cell Longev. 2016, 2016, 8627384. [Google Scholar] [CrossRef] [PubMed]
- Avrahami, D.; Li, C.; Zhang, J.; Schug, J.; Avrahami, R.; Rao, S.; Stadler, M.B.; Burger, L.; Schübeler, D.; Glaser, B.; et al. Aging-Dependent Demethylation of Regulatory Elements Correlates with Chromatin State and Improved β Cell Function. Cell Metab. 2015, 22, 619–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takasugi, M.; Hayakawa, K.; Arai, D.; Shiota, K. Age- and Sex-Dependent DNA Hypomethylation Controlled by Growth Hormone in Mouse Liver. Mech. Ageing Dev. 2013, 134, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Thalheim, T.; Herberg, M.; Galle, J. Linking DNA Damage and Age-Related Promoter DNA Hyper-Methylation in the Intestine. Genes 2018, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Khalil, H.; Tazi, M.; Caution, K.; Ahmed, A.; Kanneganti, A.; Assani, K.; Kopp, B.; Marsh, C.; Dakhlallah, D.; Amer, A.O. Aging Is Associated with Hypermethylation of Autophagy Genes in Macrophages. Epigenetics 2016, 11, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Gale, C.R.; Marioni, R.E.; Harris, S.E.; Starr, J.M.; Deary, I.J. DNA Methylation and the Epigenetic Clock in Relation to Physical Frailty in Older People: The Lothian Birth Cohort 1936. Clin. Epigenet. 2018, 10, 101. [Google Scholar] [CrossRef] [PubMed]
- Ciccarone, F.; Malavolta, M.; Calabrese, R.; Guastafierro, T.; Bacalini, M.G.; Reale, A.; Franceschi, C.; Capri, M.; Hervonen, A.; Hurme, M.; et al. Age-Dependent Expression of DNMT1 and DNMT3B in PBMCs from a Large European Population Enrolled in the Mark-Age Study. Aging Cell 2016, 15, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.P. Aging and Epigenetic Drift: A. Vicious Cycle. J. Clin. Investig. 2014, 124, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.M.; Hemstedt, T.J.; Bading, H. Rescue of Aging-Associated Decline in Dnmt3a2 Expression Restores Cognitive Abilities. Nat. Neurosci. 2012, 15, 1111–1113. [Google Scholar] [CrossRef] [PubMed]
- Maegawa, S.; Hinkal, G.; Kim, H.S.; Shen, L.; Zhang, L.; Zhang, J.; Zhang, N.; Liang, S.; Donehower, L.A.; Issa, J.P. Widespread and Tissue Specific Age-Related DNA Methylation Changes in Mice. Genome Res. 2010, 20, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Bolund, A.C.S.; Starnawska, A.; Miller, M.R.; Schlunssen, V.; Backer, V.; Borglum, A.D.; Christensen, K.; Tan, Q.; Christiansen, L.; Sigsgaard, T. Lung Function Discordance in Monozygotic Twins and Associated Differences in Blood DNA Methylation. Clin. Epigenet. 2017, 9, 132. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.T.; Tsai, P.C.; Yang, T.P.; Pidsley, R.; Nisbet, J.; Glass, D.; Mangino, M.; Zhai, G.; Zhang, F.; Valdes, A.; et al. Epigenome-Wide Scans Identify Differentially Methylated Regions for Age and Age-Related Phenotypes in a Healthy Ageing Population. PLoS Genet. 2012, 8, e1002629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De F C Lichtenfels, A.J.; van der Plaat, D.A.; de Jong, K.; van Diemen, C.C.; Postma, D.S.; Nedeljkovic, I.; van Duijn, C.M.; Amin, N.; la Bastide-van Gemert, S.; de Vries, M.; et al. Long-Term Air Pollution Exposure, Genome-Wide DNA Methylation and Lung Function in the Lifelines Cohort Study. Environ. Health Perspect. 2018, 126, 027004. [Google Scholar] [CrossRef] [PubMed]
- Nwanaji-Enwerem, J.C.; Colicino, E.; Dai, L.; Cayir, A.; Sanchez-Guerra, M.; Laue, H.E.; Nguyen, V.T.; Di, Q.; Just, A.C.; Hou, L.; et al. Impacts of the Mitochondrial Genome on the Relationship of Long-Term Ambient Fine Particle Exposure with Blood DNA Methylation Age. Environ. Sci. Technol. 2017, 51, 8185–8195. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Meng, X.; Zhao, A.; Wang, C.; Yang, C.; Li, H.; Cai, J.; Zhao, Z.; Kan, H. DNA Hypomethylation and Its Mediation in the Effects of Fine Particulate Air Pollution on Cardiovascular Biomarkers: A Randomized Crossover Trial. Environ. Int. 2016, 94, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, R.; Shi, M.; Cai, J.; Shi, J.; Yang, C.; Li, H.; Lin, Z.; Meng, X.; Liu, C.; et al. Possible Mediation by Methylation in Acute Inflammation Following Personal Exposure to Fine Particulate Air Pollution. Am. J. Epidemiol. 2018, 187, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Panni, T.; Mehta, A.J.; Schwartz, J.D.; Baccarelli, A.A.; Just, A.C.; Wolf, K.; Wahl, S.; Cyrys, J.; Kunze, S.; Strauch, K.; et al. Genome-Wide Analysis of DNA Methylation and Fine Particulate Matter Air Pollution in Three Study Populations: Kora F3, Kora F4, and the Normative Aging Study. Environ. Health Perspect. 2016, 124, 983–990. [Google Scholar] [CrossRef] [PubMed]
- Ward-Caviness, C.K.; Nwanaji-Enwerem, J.C.; Wolf, K.; Wahl, S.; Colicino, E.; Trevisi, L.; Kloog, I.; Just, A.C.; Vokonas, P.; Cyrys, J.; et al. Long-Term Exposure to Air Pollution Is Associated with Biological Aging. Oncotarget 2016, 7, 74510–74525. [Google Scholar] [CrossRef] [PubMed]
- Maghbooli, Z.; Hossein-Nezhad, A.; Adabi, E.; Asadollah-Pour, E.; Sadeghi, M.; Mohammad-Nabi, S.; Zakeri Rad, L.; Malek Hosseini, A.A.; Radmehr, M.; Faghihi, F.; et al. Air Pollution During Pregnancy and Placental Adaptation in the Levels of Global DNA Methylation. PLoS ONE 2018, 13, e0199772. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.F.; Verkaik-Schakel, R.N.; Timens, W.; Kobzik, L.; Plosch, T.; Hylkema, M.N. The Fetal Programming Effect of Prenatal Smoking on IGF1R and IGF1 Methylation Is Organ- and Sex-Specific. Epigenetics 2017, 12, 1076–1091. [Google Scholar] [CrossRef] [PubMed]
- Drake, A.J.; O’Shaughnessy, P.J.; Bhattacharya, S.; Monteiro, A.; Kerrigan, D.; Goetz, S.; Raab, A.; Rhind, S.M.; Sinclair, K.D.; Meharg, A.A.; et al. In Utero Exposure to Cigarette Chemicals Induces Sex-Specific Disruption of One-Carbon Metabolism and DNA Methylation in the Human Fetal Liver. BMC Med. 2015, 13, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, M.; Fink, B.; Thurmann, L.; Eszlinger, M.; Herberth, G.; Lehmann, I. Tobacco Smoking Differently Influences Cell Types of the Innate and Adaptive Immune System-Indications from Cpg Site Methylation. Clin. Epigenet. 2015, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, H.M.; Wang, W.; Sen, S.; Destefano Shields, C.; Lee, S.S.; Zhang, Y.W.; Clements, E.G.; Cai, Y.; van Neste, L.; Easwaran, H.; et al. Oxidative Damage Targets Complexes Containing DNA Methyltransferases, Sirt1, and Polycomb Members to Promoter Cpg Islands. Cancer Cell 2011, 20, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Vaz, M.; Hwang, S.Y.; Kagiampakis, I.; Phallen, J.; Patil, A.; O’Hagan, H.M.; Murphy, L.; Zahnow, C.A.; Gabrielson, E.; Velculescu, V.E.; et al. Chronic Cigarette Smoke-Induced Epigenomic Changes Precede Sensitization of Bronchial Epithelial Cells to Single-Step Transformation by Kras Mutations. Cancer Cell 2017, 32, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Elgizouli, M.; Schottker, B.; Holleczek, B.; Nieters, A.; Brenner, H. Smoking-Associated DNA Methylation Markers Predict Lung Cancer Incidence. Clin. Epigenet. 2016, 8, 127. [Google Scholar] [CrossRef] [PubMed]
- Steenaard, R.V.; Ligthart, S.; Stolk, L.; Peters, M.J.; van Meurs, J.B.; Uitterlinden, A.G.; Hofman, A.; Franco, O.H.; Dehghan, A. Tobacco Smoking Is Associated with Methylation of Genes Related to Coronary Artery Disease. Clin. Epigenet. 2015, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, Y.; Saum, K.U.; Schottker, B.; Breitling, L.P.; Brenner, H. Tobacco Smoking and Smoking-Related DNA Methylation Are Associated with the Development of Frailty among Older Adults. Epigenetics 2017, 12, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, Y.; Breitling, L.P.; Brenner, H. Relationship of Tobacco Smoking and Smoking-Related DNA Methylation with Epigenetic Age Acceleration. Oncotarget 2016, 7, 46878–46889. [Google Scholar] [CrossRef] [PubMed]
- Ligthart, S.; Steenaard, R.V.; Peters, M.J.; van Meurs, J.B.; Sijbrands, E.; Uitterlinden, A.G.; Bonder, M.J.; BIOS Consortium; Hofman, A.; Franco, O.H.; et al. Tobacco Smoking Is Associated with DNA Methylation of Diabetes Susceptibility Genes. Diabetologia 2016, 59, 998–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Marioni, R.E.; Hedman, Å.K.; Pfeiffer, L.; Tsai, P.C.; Reynolds, L.M.; Just, A.C.; Duan, Q.; Boer, C.G.; Tanaka, T.; et al. A DNA Methylation Biomarker of Alcohol Consumption. Mol. Psychiatry 2018, 23, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Sharp, G.C.; Arathimos, R.; Reese, S.E.; Page, C.M.; Felix, J.; Küpers, L.K.; Rifas-Shiman, S.L.; Liu, C. Cohorts for Heart and Aging Research in Genomic Epidemiology plus (CHARGE+) Methylation Alcohol Working Group; Burrows, K.; et al. Maternal Alcohol Consumption and Offspring DNA Methylation: Findings from Six General Population-Based Birth Cohorts. Epigenomics 2018, 10, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Denham, J.; O’Brien, B.J.; Harvey, J.T.; Charchar, F.J. Genome-Wide Sperm DNA Methylation Changes after 3 Months of Exercise Training in Humans. Epigenomics 2015, 7, 717–731. [Google Scholar] [CrossRef] [PubMed]
- Barres, R.; Yan, J.; Egan, B.; Treebak, J.T.; Rasmussen, M.; Fritz, T.; Caidahl, K.; Krook, A.; O’Gorman, D.J.; Zierath, J.R. Acute Exercise Remodels Promoter Methylation in Human Skeletal Muscle. Cell Metab. 2012, 15, 405–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maejima, H.; Kanemura, N.; Kokubun, T.; Murata, K.; Takayanagi, K. Exercise Enhances Cognitive Function and Neurotrophin Expression in the Hippocampus Accompanied by Changes in Epigenetic Programming in Senescence-Accelerated Mice. Neurosci. Lett. 2018, 665, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Ronn, T.; Volkov, P.; Davegårdh, C.; Dayeh, T.; Hall, E.; Olsson, A.H.; Nilsson, E.; Tornberg, A.; Dekker Nitert, M.; Eriksson, K.F.; et al. A Six Months Exercise Intervention Influences the Genome-Wide DNA Methylation Pattern in Human Adipose Tissue. PLoS Genet. 2013, 9, e1003572. [Google Scholar] [CrossRef] [PubMed]
- Barres, R.; Kirchner, H.; Rasmussen, M.; Yan, J.; Kantor, F.R.; Krook, A.; Naslund, E.; Zierath, J.R. Weight Loss after Gastric Bypass Surgery in Human Obesity Remodels Promoter Methylation. Cell Rep. 2013, 3, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Hahn, O.; Grönke, S.; Stubbs, T.M.; Ficz, G.; Hendrich, O.; Krueger, F.; Andrews, S.; Zhang, Q.; Wakelam, M.J.; Beyer, A.; et al. Dietary Restriction Protects from Age-Associated DNA Methylation and Induces Epigenetic Reprogramming of Lipid Metabolism. Genome Biol. 2017, 18, 56. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.J.; Robertson, N.A.; Rather, M.I.; Thomson, J.P.; McBryan, T.; Sproul, D.; Wang, T.; Brock, C.; Clark, W.; Ideker, T.; et al. Diverse Interventions That Extend Mouse Lifespan Suppress Shared Age-Associated Epigenetic Changes at Critical Gene Regulatory Regions. Genome Biol. 2017, 18, 58. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.H.; Lee, E.K.; Choi, Y.J.; An, H.J.; Jeong, H.O.; Park, D.; Kim, B.C.; Yu, B.P.; Bhak, J.; Chung, H.Y. Short-Term Calorie Restriction Ameliorates Genomewide, Age-Related Alterations in DNA Methylation. Aging Cell 2016, 15, 1074–1081. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, R.; Nwanaji-Enwerem, J.C.; Samet, M.; Ward-Caviness, C.K. DNA Methylation Age-Environmental Influences, Health Impacts, and Its Role in Environmental Epidemiology. Curr. Environ. Health Rep. 2018, 5, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Austin, M.K.; Chen, E.; Ross, K.M.; McEwen, L.M.; Maclsaac, J.L.; Kobor, M.S.; Miller, G.E. Early-Life Socioeconomic Disadvantage, Not Current, Predicts Accelerated Epigenetic Aging of Monocytes. Psychoneuroendocrinology 2018, 97, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, X.; Yu, K.; Jiang, H.; Zhang, Y.; Wang, B.; Liu, X.; Deng, S.; Hu, J.; Deng, Q.; et al. Exposure to Polycyclic Aromatic Hydrocarbons and Accelerated DNA Methylation Aging. Environ. Health Perspect. 2018, 126, 067005. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.B.; Fabris, V.; Lecke, S.B.; Maturana, M.A.; Spritzer, P.M. Association between Global Leukocyte DNA Methylation and Cardiovascular Risk in Postmenopausal Women. BMC Med. Genet. 2016, 17, 71. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Guo, Z.; Guo, Y.; Li, M.; Yan, H.; Cheng, J.; Wang, C.; Hong, G. Common DNA Methylation Alterations of Alzheimer’s Disease and Aging in Peripheral Whole Blood. Oncotarget 2016, 7, 19089–19098. [Google Scholar] [PubMed]
- Marioni, R.E.; Shah, S.; McRae, A.F.; Chen, B.H.; Colicino, E.; Harris, S.E.; Gibson, J.; Henders, A.K.; Redmond, P.; Cox, S.R.; et al. DNA Methylation Age of Blood Predicts All-Cause Mortality in Later Life. Genome Biol. 2015, 16, 25. [Google Scholar] [CrossRef] [PubMed]
- Watson, C.T.; Roussos, P.; Garg, P.; Ho, D.J.; Azam, N.; Katsel, P.L.; Haroutunian, V.; Sharp, A.J. Genome-Wide DNA Methylation Profiling in the Superior Temporal Gyrus Reveals Epigenetic Signatures Associated with Alzheimer’s Disease. Genome Med. 2016, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA Methylation Age of Human Tissues and Cell Types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Cruickshanks, H.A.; McBryan, T.; Nelson, D.M.; Vanderkraats, N.D.; Shah, P.P.; van Tuyn, J.; Singh Rai, T.; Brock, C.; Donahue, G.; Dunican, D.S.; et al. Senescent Cells Harbour Features of the Cancer Epigenome. Nat. Cell Biol 2013, 15, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Kresovich, J.K.; Joyce, B.T.; Gao, T.; Zheng, Y.; Zhang, Z.; Achenbach, C.J.; Murphy, R.L.; Just, A.C.; Shen, J.; Yang, H.; et al. Promoter Methylation of Pgc1a and Pgc1b Predicts Cancer Incidence in a Veteran Cohort. Epigenomics 2018, 10, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Perna, L.; Zhang, Y.; Mons, U.; Holleczek, B.; Saum, K.U.; Brenner, H. Epigenetic Age Acceleration Predicts Cancer, Cardiovascular, and All-Cause Mortality in a German Case Cohort. Clin. Epigenet. 2016, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Meliso, F.M.; Micali, D.; Silva, C.T.; Sabedot, T.S.; Coetzee, S.G.; Koch, A.; Fahlbusch, F.B.; Noushmehr, H.; Schneider-Stock, R.; Jasiulionis, M.G. Sirt1 Regulates Mxd1 During Malignant Melanoma Progression. Oncotarget 2017, 8, 114540–114553. [Google Scholar] [CrossRef] [PubMed]
- Joyce, B.T.; Gao, T.; Zheng, Y.; Liu, L.; Zhang, W.; Dai, Q.; Shrubsole, M.J.; Hibler, E.A.; Cristofanilli, M.; Zhang, H.; et al. Prospective Changes in Global DNA Methylation and Cancer Incidence and Mortality. Br. J. Cancer 2016, 115, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Nishida, N.; Iwanishi, M.; Minami, T.; Chishina, H.; Arizumi, T.; Takita, M.; Kitai, S.; Yada, N.; Ida, H.; Hagiwara, S.; et al. Hepatic DNA Methylation Is Affected by Hepatocellular Carcinoma Risk in Patients with and without Hepatitis Virus. Dig. Dis. 2015, 33, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Ianov, L.; Riva, A.; Kumar, A.; Foster, T.C. DNA Methylation of Synaptic Genes in the Prefrontal Cortex Is Associated with Aging and Age-Related Cognitive Impairment. Front. Aging Neurosci. 2017, 9, 249. [Google Scholar] [CrossRef] [PubMed]
- Spiers, H.; Hannon, E.; Wells, S.; Williams, B.; Fernandes, C.; Mill, J. Age-Associated Changes in DNA Methylation across Multiple Tissues in an Inbred Mouse Model. Mech. Ageing Dev. 2016, 154, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Kananen, L.; Marttila, S.; Nevalainen, T.; Jylhava, J.; Mononen, N.; Kahonen, M.; Raitakari, O.T.; Lehtimaki, T.; Hurme, M. Aging-Associated DNA Methylation Changes in Middle-Aged Individuals: The Young Finns Study. BMC Genom. 2016, 17, 103. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Jiang, Z.; Xia, Y.; Lou, P.; Chen, L.; Wang, H.; Bai, L.; Xie, Y.; Liu, Y.; Li, W.; et al. Genome-Wide DNA Methylation Changes in Skeletal Muscle between Young and Middle-Aged Pigs. BMC Genom. 2014, 15, 653. [Google Scholar] [CrossRef] [PubMed]
- Svane, A.M.; Soerensen, M.; Lund, J.; Tan, Q.; Jylhävä, J.; Wang, Y.; Pedersen, N.L.; Hägg, S.; Debrabant, B.; Deary, I.J.; et al. DNA Methylation and All-Cause Mortality in Middle-Aged and Elderly Danish Twins. Genes 2018, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y.; et al. An Epigenetic Biomarker of Aging for Lifespan and Healthspan. Aging 2018, 10, 573–591. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.H.; Marioni, R.E.; Colicino, E.; Peters, M.J.; Ward-Caviness, C.K.; Tsai, P.C.; Roetker, N.S.; Just, A.C.; Demerath, E.W.; Guan, W.; et al. DNA Methylation-Based Measures of Biological Age: Meta-Analysis Predicting Time to Death. Aging 2016, 8, 1844–1865. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, L.; Lenart, A.; Tan, Q.; Vaupel, J.W.; Aviv, A.; McGue, M.; Christensen, K. DNA Methylation Age Is Associated with Mortality in a Longitudinal Danish Twin Study. Aging Cell 2016, 15, 149–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Q.; Weidner, C.I.; Costa, I.G.; Marioni, R.E.; Ferreira, M.R.; Deary, I.J.; Wagner, W. DNA Methylation Levels at Individual Age-Associated Cpg Sites Can Be Indicative for Life Expectancy. Aging 2016, 8, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Niu, Z.; Chen, Y.; Tu, Q.; Zhang, Y.; Chen, W.; Tong, W.; Zhang, Z. Repetitive Element DNA Methylation Is Associated with Menopausal Age. Aging Dis. 2018, 9, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-Wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Ponnaluri, V.K.C.; Esteve, P.O.; Ruse, C.I.; Pradhan, S. S-Adenosylhomocysteine Hydrolase Participates in DNA Methylation Inheritance. J. Mol. Biol. 2018, 430, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- Esteve, P.O.; Terragni, J.; Deepti, K.; Chin, H.G.; Dai, N.; Espejo, A.; Correa, I.R., Jr.; Bedford, M.T.; Pradhan, S. Methyllysine Reader Plant Homeodomain (Phd) Finger Protein 20-Like 1 (Phf20l1) Antagonizes DNA (Cytosine-5) Methyltransferase 1 (DNMT1) Proteasomal Degradation. J. Biol. Chem. 2014, 289, 8277–8287. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.M.; Griffiths, A.D.; Tawfik, D.S.; Loakes, D. Determinants of Cofactor Binding to DNA Methyltransferases: Insights from a Systematic Series of Structural Variants of S-Adenosylhomocysteine. Org. Biomol. Chem. 2005, 3, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Denomme, M.M.; White, C.R.; Leung, K.Y.; Lee, M.B.; Greene, N.D.; Mann, M.R.; Trasler, J.M.; Baltz, J.M. Both the Folate Cycle and Betaine-Homocysteine Methyltransferase Contribute Methyl Groups for DNA Methylation in Mouse Blastocysts. FASEB J. 2015, 29, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Frostesjo, L.; Holm, I.; Grahn, B.; Page, A.W.; Bestor, T.H.; Heby, O. Interference with DNA Methyltransferase Activity and Genome Methylation During F9 Teratocarcinoma Stem Cell Differentiation Induced by Polyamine Depletion. J. Biol. Chem. 1997, 272, 4359–4366. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, H.; Zhou, D.; Moody, L.; Lezmi, S.; Chen, H.; Pan, Y.X. High-Fat Diet Caused Widespread Epigenomic Differences on Hepatic Methylome in Rat. Physiol. Genom. 2015, 47, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Altobelli, G.; Bogdarina, I.G.; Stupka, E.; Clark, A.J.; Langley-Evans, S. Genome-Wide Methylation and Gene Expression Changes in Newborn Rats Following Maternal Protein Restriction and Reversal by Folic Acid. PLoS ONE 2013, 8, e82989. [Google Scholar] [CrossRef] [PubMed]
- Yoon, A.; Tammen, S.A.; Park, S.; Han, S.N.; Choi, S.W. Genome-Wide Hepatic DNA Methylation Changes in High-Fat Diet-Induced Obese Mice. Nutr. Res. Pract. 2017, 11, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Tammen, S.A.; Liu, Z.; Friso, S. A Lifelong Exposure to a Western-Style Diet, but Not Aging, Alters Global DNA Methylation in Mouse Colon. Nutr. Res. Pract. 2015, 9, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Van Straten, E.M.; Bloks, V.W.; Huijkman, N.C.; Baller, J.F.; van Meer, H.; Lutjohann, D.; Kuipers, F.; Plosch, T. The Liver X-Receptor Gene Promoter Is Hypermethylated in a Mouse Model of Prenatal Protein Restriction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 298, R275–R282. [Google Scholar] [CrossRef] [PubMed]
- Kamat, P.K.; Vacek, J.C.; Kalani, A.; Tyagi, N. Homocysteine Induced Cerebrovascular Dysfunction: A Link to Alzheimer’s Disease Etiology. Open Neurol. J. 2015, 9, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.C.; Lin, Y.Y.; Liu, X.H.; Zhao, Y.C.; Ma, X.Y.; Yu, J.; Liu, X.Y.; Zhao, Y.X. Homocysteine Is Associated with Exaggerated Morning Blood Pressure Surge in Patients with Acute Ischemic Stroke. J. Stroke Cerebrovasc. Dis. 2018, 27, 2650–2656. [Google Scholar] [CrossRef] [PubMed]
- Snyder, H.M.; Corriveau, R.A.; Craft, S.; Faber, J.E.; Greenberg, S.M.; Knopman, D.; Lamb, B.T.; Montine, T.J.; Nedergaard, M.; Schaffer, C.B.; et al. Vascular Contributions to Cognitive Impairment and Dementia Including Alzheimer’s Disease. Alzheimers Dement. 2015, 11, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Okura, T.; Miyoshi, K.; Irita, J.; Enomoto, D.; Nagao, T.; Kukida, M.; Tanino, A.; Kudo, K.; Pei, Z.; Higaki, J. Hyperhomocysteinemia Is One of the Risk Factors Associated with Cerebrovascular Stiffness in Hypertensive Patients, Especially Elderly Males. Sci. Rep. 2014, 4, 5663. [Google Scholar] [CrossRef] [PubMed]
- Shah, H.; Jan, M.U.; Altaf, A.; Salahudin, M. Correlation of Hyper-Homocysteinemia with Coronary Artery Disease in Absence of Conventional Risk Factors among Young Adults. J. Saudi. Heart Assoc. 2018, 30, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Catena, C.; Colussi, G.; Nait, F.; Capobianco, F.; Sechi, L.A. Elevated Homocysteine Levels Are Associated with the Metabolic Syndrome and Cardiovascular Events in Hypertensive Patients. Am. J. Hypertens. 2015, 28, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Catena, C.; Colussi, G.; Url-Michitsch, M.; Nait, F.; Sechi, L.A. Subclinical Carotid Artery Disease and Plasma Homocysteine Levels in Patients with Hypertension. J. Am. Soc. Hypertens. 2015, 9, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Lee, M.H.; Fujii, T.; Fujii, N.; Moon, Y. Association of the Urine Homocysteine/Creatinine Ratio to Proinflammatory Cytokine, Natural Anticoagulant, and Nitric Oxide Levels in Cerebrovascular Disease. Ann. Clin. Lab. Sci. 2014, 44, 461–465. [Google Scholar] [PubMed]
- Schaffer, A.; Verdoia, M.; Cassetti, E.; Marino, P.; Suryapranata, H.; De Luca, G.; Novara Atherosclerosis Study, G. Relationship between Homocysteine and Coronary Artery Disease. Results from a Large Prospective Cohort Study. Thromb. Res. 2014, 134, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Miwa, K.; Tanaka, M.; Okazaki, S.; Yagita, Y.; Sakaguchi, M.; Mochizuki, H.; Kitagawa, K. Increased Total Homocysteine Levels Predict the Risk of Incident Dementia Independent of Cerebral Small-Vessel Diseases and Vascular Risk Factors. J. Alzheimers Dis. 2016, 49, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Jadavji, N.M.; Farr, T.D.; Lips, J.; Khalil, A.A.; Boehm-Sturm, P.; Foddis, M.; Harms, C.; Fuchtemeier, M.; Dirnagl, U. Elevated Levels of Plasma Homocysteine, Deficiencies in Dietary Folic Acid and Uracil-DNA Glycosylase Impair Learning in a Mouse Model of Vascular Cognitive Impairment. Behav. Brain Res. 2015, 283, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Kamat, P.K.; Kyles, P.; Kalani, A.; Tyagi, N. Hydrogen Sulfide Ameliorates Homocysteine-Induced Alzheimer’s Disease-Like Pathology, Blood-Brain Barrier Disruption, and Synaptic Disorder. Mol. Neurobiol. 2016, 53, 2451–2467. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Sanchez, N.; Alvarez-Rios, A.I.; Guerrero, J.M.; Garcia-Garcia, F.J.; Rodriguez-Manas, L.; Cruz-Chamorro, I.; Lardone, P.J.; Carrillo-Vico, A. Homocysteine Levels Are Associated with Bone Resorption in Pre-Frail and Frail Spanish Women: The Toledo Study for Healthy Aging. Exp. Gerontol. 2018, 108, 201–208. [Google Scholar] [CrossRef] [PubMed]
- McLean, R.R.; Jacques, P.F.; Selhub, J.; Tucker, K.L.; Samelson, E.J.; Broe, K.E.; Hannan, M.T.; Cupples, L.A.; Kiel, D.P. Homocysteine as a Predictive Factor for Hip Fracture in Older Persons. N. Engl. J. Med. 2004, 350, 2042–2049. [Google Scholar] [CrossRef] [PubMed]
- Van Meurs, J.B.; Dhonukshe-Rutten, R.A.; Pluijm, S.M.; van der Klift, M.; de Jonge, R.; Lindemans, J.; de Groot, L.C.; Hofman, A.; Witteman, J.C.; van Leeuwen, J.P.; et al. Homocysteine Levels and the Risk of Osteoporotic Fracture. N. Engl. J. Med. 2004, 350, 2033–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Z.; Guan, Y.; Huo, Y.R.; Liu, S.; Zhang, M.; Lu, H.; Yue, W.; Wang, J.; Ji, Y. Elevated Total Homocysteine Levels in Acute Ischemic Stroke Are Associated with Long-Term Mortality. Stroke 2015, 46, 2419–2425. [Google Scholar] [CrossRef] [PubMed]
- Shankle, W.R.; Hara, J.; Barrentine, L.W.; Curole, M.V. Cerefolinnac Therapy of Hyperhomocysteinemia Delays Cortical and White Matter Atrophy in Alzheimer’s Disease and Cerebrovascular Disease. J. Alzheimers Dis. 2016, 54, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.L.; Bowe, S.J.; Crowe, T.C. A Combination of Omega-3 Fatty Acids, Folic Acid and B-Group Vitamins Is Superior at Lowering Homocysteine Than Omega-3 Alone: A Meta-Analysis. Nutr. Res. 2016, 36, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Mazza, A.; Cicero, A.F.; Ramazzina, E.; Lenti, S.; Schiavon, L.; Casiglia, E.; Gussoni, G. Nutraceutical Approaches to Homocysteine Lowering in Hypertensive Subjects at Low Cardiovascular Risk: A Multicenter, Randomized Clinical Trial. J. Biol. Regul. Homeost. Agents 2016, 30, 921–927. [Google Scholar] [PubMed]
- Toole, J.F.; Malinow, M.R.; Chambless, L.E.; Spence, J.D.; Pettigrew, L.C.; Howard, V.J.; Sides, E.G.; Wang, C.H.; Stampfer, M. Lowering Homocysteine in Patients with Ischemic Stroke to Prevent Recurrent Stroke, Myocardial Infarction, and Death: The Vitamin Intervention for Stroke Prevention (Visp) Randomized Controlled Trial. JAMA 2004, 291, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine (SEARCH) Collaborative Group; Armitage, J.M.; Bowman, L.; Clarke, R.J.; Wallendszus, K.; Bulbulia, R.; Rahimi, K.; Haynes, R.; Parish, S.; Sleight, P.; et al. Effects of Homocysteine-Lowering with Folic Acid Plus Vitamin B12 Vs Placebo on Mortality and Major Morbidity in Myocardial Infarction Survivors: A Randomized Trial. JAMA 2010, 303, 2486–2494. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Li, W.; Wang, P.; Lv, X.; Gao, Y.; Huang, G. Folic Acid Inhibits Homocysteine-Induced Cell Apoptosis in Human Umbilical Vein Endothelial Cells. Mol. Cell Biochem. 2018, 444, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Saposnik, G.; Ovbiagele, B.; Markovic, D.; Towfighi, A. Effect of B-Vitamins on Stroke Risk among Individuals with Vascular Disease Who Are Not on Antiplatelets: A Meta-Analysis. Int. J. Stroke 2016, 11, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Li, W.; Lv, X.; Wang, P.; Gao, Y.; Huang, G. Folic Acid Supplementation Delays Atherosclerotic Lesion Development by Modulating Mcp1 and Vegf DNA Methylation Levels in Vivo and in Vitro. Int. J. Mol. Sci. 2017, 18, 990. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Xu, C.H.; Xu, Y.N.; Wang, Y.L.; Wang, M. The Effect of Folate Fortification on Folic Acid-Based Homocysteine-Lowering Intervention and Stroke Risk: A Meta-Analysis. Public Health Nutr. 2015, 18, 1514–1521. [Google Scholar] [CrossRef] [PubMed]
- VITATOPS Trial Study Group. B Vitamins in Patients with Recent Transient Ischaemic Attack or Stroke in the Vitamins to Prevent Stroke (Vitatops) Trial: A Randomised, Double-Blind, Parallel, Placebo-Controlled Trial. Lancet Neurol. 2010, 9, 855–865. [Google Scholar] [CrossRef] [Green Version]
- Nigwekar, S.U.; Kang, A.; Zoungas, S.; Cass, A.; Gallagher, M.P.; Kulshrestha, S.; Navaneethan, S.D.; Perkovic, V.; Strippoli, G.F.; Jardine, M.J. Interventions for Lowering Plasma Homocysteine Levels in Dialysis Patients. Cochrane Database Syst. Rev. 2016, CD004683. [Google Scholar] [CrossRef]
- Marti-Carvajal, A.J.; Sola, I.; Lathyris, D.; Dayer, M. Homocysteine-Lowering Interventions for Preventing Cardiovascular Events. Cochrane Database Syst. Rev. 2017, 8, CD006612. [Google Scholar] [PubMed]
- Clarke, R.; Bennett, D.; Parish, S.; Lewington, S.; Skeaff, M.; Eussen, S.J.; Lewerin, C.; Stott, D.J.; Armitage, J.; Hankey, G.J.; et al. Effects of Homocysteine Lowering with B Vitamins on Cognitive Aging: Meta-Analysis of 11 Trials with Cognitive Data on 22,000 Individuals. Am. J. Clin. Nutr. 2014, 100, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Lonn, E.; Yusuf, S.; Arnold, M.J.; Sheridan, P.; Pogue, J.; Micks, M.; McQueen, M.J.; Probstfield, J.; Fodor, G.; Held, C.; et al. Homocysteine Lowering with Folic Acid and B Vitamins in Vascular Disease. N. Engl. J. Med. 2006, 354, 1567–1577. [Google Scholar] [PubMed]
- Stott, D.J.; MacIntosh, G.; Lowe, G.D.; Rumley, A.; McMahon, A.D.; Langhorne, P.; Tait, R.C.; O’Reilly, D.S.; Spilg, E.G.; MacDonald, J.B.; et al. Randomized Controlled Trial of Homocysteine-Lowering Vitamin Treatment in Elderly Patients with Vascular Disease. Am. J. Clin. Nutr. 2005, 82, 1320–1326. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Cook, N.; Manson, J.; Buring, J.E.; Albert, C.M.; Grodstein, F. A Trial of B Vitamins and Cognitive Function among Women at High Risk of Cardiovascular Disease. Am. J. Clin. Nutr. 2008, 88, 1602–1610. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, V.A.; Kesse-Guyot, E.; Barberger-Gateau, P.; Fezeu, L.; Hercberg, S.; Galan, P. Cognitive Function after Supplementation with B Vitamins and Long-Chain Omega-3 Fatty Acids: Ancillary Findings from the Su.Fol.Om3 Randomized Trial. Am. J. Clin. Nutr. 2011, 94, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Stone, K.L.; Lui, L.Y.; Christen, W.G.; Troen, A.M.; Bauer, D.C.; Kado, D.; Schambach, C.; Cummings, S.R.; Manson, J.E. Effect of Combination Folic Acid, Vitamin B6, and Vitamin B12 Supplementation on Fracture Risk in Women: A Randomized, Controlled Trial. J. Bone Miner. Res. 2017, 32, 2331–2338. [Google Scholar] [CrossRef] [PubMed]
- Garcia Lopez, M.; Bonaa, K.H.; Ebbing, M.; Eriksen, E.F.; Gjesdal, C.G.; Nygard, O.; Tell, G.S.; Ueland, P.M.; Meyer, H.E. B Vitamins and Hip Fracture: Secondary Analyses and Extended Follow-up of Two Large Randomized Controlled Trials. J. Bone Miner. Res. 2017, 32, 1981–1989. [Google Scholar] [CrossRef] [PubMed]
- Ntaios, G.; Savopoulos, C.; Karamitsos, D.; Economou, I.; Destanis, E.; Chryssogonidis, I.; Pidonia, I.; Zebekakis, P.; Polatides, C.; Sion, M.; et al. The Effect of Folic Acid Supplementation on Carotid Intima-Media Thickness in Patients with Cardiovascular Risk: A Randomized, Placebo-Controlled Trial. Int. J. Cardiol. 2010, 143, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.; Sanchez-Roman, I.; Gomez, A.; Sanchez, C.; Suarez, H.; Lopez-Torres, M.; Barja, G. Methionine and Homocysteine Modulate the Rate of Ros Generation of Isolated Mitochondria in vitro. J. Bioenergy Biomembr. 2011, 43, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yang, H.M.; Gong, D.Q.; Rose, S.P.; Pirgozliev, V.; Chen, X.S.; Wang, Z.Y. Transcriptome Analysis of Hepatic Gene Expression and DNA Methylation in Methionine- and Betaine-Supplemented Geese (Anser Cygnoides Domesticus). Poult. Sci. 2018, 97, 3463–3477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.P.; Wang, Y.H.; Ma, S.C.; Zhang, H.; Yang, A.N.; Yang, X.L.; Zhang, M.H.; Sun, J.M.; Hao, Y.J.; Jiang, Y.D. Homocysteine Inhibits Endothelial Progenitor Cells Proliferation Via DNMT1-Mediated Hypomethylation of Cyclin A. Exp. Cell Res. 2018, 362, 217–226. [Google Scholar] [CrossRef] [PubMed]
- McNeil, C.J.; Beattie, J.H.; Gordon, M.J.; Pirie, L.P.; Duthie, S.J. Differential Effects of Nutritional Folic Acid Deficiency and Moderate Hyperhomocysteinemia on Aortic Plaque Formation and Genome-Wide DNA Methylation in Vascular Tissue from Apoe−/− Mice. Clin. Epigenet. 2011, 2, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Dawson, H.; Collins, G.; Pyle, R.; Deep-Dixit, V.; Taub, D.D. The Immunoregulatory Effects of Homocysteine and Its Intermediates on T-Lymphocyte Function. Mech. Ageing Dev. 2004, 125, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Elmasry, K.; Mohamed, R.; Sharma, I.; Elsherbiny, N.M.; Liu, Y.; Al-Shabrawey, M.; Tawfik, A. Epigenetic Modifications in Hyperhomocysteinemia: Potential Role in Diabetic Retinopathy and Age-Related Macular Degeneration. Oncotarget 2018, 9, 12562–12590. [Google Scholar] [CrossRef] [PubMed]
- Badiga, S.; Siddiqui, N.R.; Macaluso, M.; Johanning, G.L.; Piyathilake, C.J. Homocysteinemia Is Associated with a Lower Degree of Pbmc Line-1 Methylation and a Higher Risk of Cin 2c in the U.S. Post-Folic Acid Fortification Era. Nutr. Cancer 2016, 68, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Li, J.G.; Barrero, C.; Gupta, S.; Kruger, W.D.; Merali, S.; Pratico, D. Homocysteine Modulates 5-Lipoxygenase Expression Level Via DNA Methylation. Aging Cell 2017, 16, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Pushpakumar, S.; Kundu, S.; Narayanan, N.; Sen, U. DNA Hypermethylation in Hyperhomocysteinemia Contributes to Abnormal Extracellular Matrix Metabolism in the Kidney. FASEB J. 2015, 29, 4713–4725. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Jiao, Y.; Yang, S.; Deng, M.; Yang, X.; Mao, C.; Sun, Y.; Ding, N.; Li, N.; Zhang, M.; et al. Homocysteine Activates Autophagy by Inhibition of Cftr Expression Via Interaction between DNA Methylation and H3k27me3 in Mouse Liver. Cell Death Dis. 2018, 9, 169. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Sun, Y.; Gao, Y.; Yang, S.; Mao, C.; Ding, N.; Deng, M.; Wang, Y.; Yang, X.; Jia, Y.; et al. Reciprocal Regulation between Mir-148a/152 and DNA Methyltransferase 1 Is Associated with Hyperhomocysteinemia-Accelerated Atherosclerosis. DNA Cell Biol. 2017, 36, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, J.A.; Dugué, P.A.; Bassett, J.K.; Hodge, A.M.; Brinkman, M.T.; Joo, J.E.; Jung, C.H.; Makalic, E.F.; Schmidt, D.; Hopper, J.L.; et al. Dietary Intake of One-Carbon Metabolism Nutrients and DNA Methylation in Peripheral Blood. Am. J. Clin. Nutr. 2018. [Google Scholar] [CrossRef] [PubMed]
- Craig, P.M.; Moon, T.W. Methionine Restriction Affects the Phenotypic and Transcriptional Response of Rainbow Trout (Oncorhynchus Mykiss) to Carbohydrate-Enriched Diets. Br. J. Nutr. 2013, 109, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Takumi, S.; Okamura, K.; Yanagisawa, H.; Sano, T.; Kobayashi, Y.; Nohara, K. The Effect of a Methyl-Deficient Diet on the Global DNA Methylation and the DNA Methylation Regulatory Pathways. J. Appl. Toxicol. 2015, 35, 1550–1556. [Google Scholar] [CrossRef] [PubMed]
- Koz, S.T.; Etem, E.O.; Baydas, G.; Yuce, H.; Ozercan, H.I.; Kuloglu, T.; Koz, S.; Etem, A.; Demir, N. Effects of Resveratrol on Blood Homocysteine Level, on Homocysteine Induced Oxidative Stress, Apoptosis and Cognitive Dysfunctions in Rats. Brain Res. 2012, 1484, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.C.; Zhang, H.P.; Jiao, Y.; Wang, Y.H.; Zhang, H.; Yang, X.L.; Yang, A.N.; Jiang, Y.D. Homocysteine-Induced Proliferation of Vascular Smooth Muscle Cells Occurs Via Pten Hypermethylation and Is Mitigated by Resveratrol. Mol. Med. Rep. 2018, 17, 5312–5319. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, A.; Barchitta, M.; Mazzone, M.G.; Giuliano, F.; Basile, G.; Agodi, A. Resveratrol Modulates Sirt1 and Dnmt Functions and Restores Line-1 Methylation Levels in Arpe-19 Cells under Oxidative Stress and Inflammation. Int. J. Mol. Sci. 2018, 19, 2118. [Google Scholar] [CrossRef] [PubMed]
- Medina-Aguilar, R.; Pérez-Plasencia, C.; Marchat, L.A.; Gariglio, P.; García Mena, J.; Rodríguez Cuevas, S.; Ruíz-García, E.; Astudillo-de la Vega, H.; Hernández Juárez, J.; Flores-Pérez, A.; et al. Methylation Landscape of Human Breast Cancer Cells in Response to Dietary Compound Resveratrol. PLoS ONE 2016, 11, e0157866. [Google Scholar] [CrossRef] [PubMed]
- Zanatta, A.; Cecatto, C.; Ribeiro, R.T.; Amaral, A.U.; Wyse, A.T.; Leipnitz, G.; Wajner, M. S-Adenosylmethionine Promotes Oxidative Stress and Decreases Na(+), K(+)-Atpase Activity in Cerebral Cortex Supernatants of Adolescent Rats: Implications for the Pathogenesis of S-Adenosylhomocysteine Hydrolase Deficiency. Mol. Neurobiol. 2018, 55, 5868–5878. [Google Scholar] [CrossRef] [PubMed]
- Sanz, A.; Caro, P.; Ayala, V.; Portero-Otin, M.; Pamplona, R.; Barja, G. Methionine Restriction Decreases Mitochondrial Oxygen Radical Generation and Leak as Well as Oxidative Damage to Mitochondrial DNA and Proteins. FASEB J. 2006, 20, 1064–1073. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.; Caro, P.; Sanchez, I.; Naudi, A.; Jove, M.; Portero-Otin, M.; Lopez-Torres, M.; Pamplona, R.; Barja, G. Effect of Methionine Dietary Supplementation on Mitochondrial Oxygen Radical Generation and Oxidative DNA Damage in Rat Liver and Heart. J. Bioenerg. Biomembr. 2009, 41, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.N.; Zhang, H.P.; Sun, Y.; Yang, X.L.; Wang, N.; Zhu, G.; Zhang, H.; Xu, H.; Ma, S.C.; Zhang, Y.; et al. High-Methionine Diets Accelerate Atherosclerosis by Hhcy-Mediated Fabp4 Gene Demethylation Pathway Via DNMT1 in ApoE(−/−) Mice. FEBS Lett. 2015, 589, 3998–4009. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Zhang, H.; Ying, Z.; Li, Y.; Zhou, L.; Wang, F.; Zhang, L.; Wang, T. Effects of Dietary L-Methionine Supplementation on Intestinal Integrity and Oxidative Status in Intrauterine Growth-Retarded Weanling Piglets. Eur. J. Nutr. 2017. [Google Scholar] [CrossRef] [PubMed]
- Aissa, A.F.; Amaral, C.L.D.; Venancio, V.P.; Machado, C.D.S.; Hernandes, L.C.; Santos, P.; Curi, R.; Bianchi, M.L.P.; Antunes, L.M.G. Methionine-Supplemented Diet Affects the Expression of Cardiovascular Disease-Related Genes and Increases Inflammatory Cytokines in Mice Heart and Liver. J. Toxicol. Environ. Health A 2017, 80, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Miousse, I.R.; Pathak, R.; Garg, S.; Skinner, C.M.; Melnyk, S.; Pavliv, O.; Hendrickson, H.; Landes, R.D.; Lumen, A.; Tackett, A.J.; et al. Short-Term Dietary Methionine Supplementation Affects One-Carbon Metabolism and DNA Methylation in the Mouse Gut and Leads to Altered Microbiome Profiles, Barrier Function, Gene Expression and Histomorphology. Genes Nutr. 2017, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Parrish, R.R.; Buckingham, S.C.; Mascia, K.L.; Johnson, J.J.; Matyjasik, M.M.; Lockhart, R.M.; Lubin, F.D. Methionine Increases Bdnf DNA Methylation and Improves Memory in Epilepsy. Ann. Clin. Transl. Neurol. 2015, 2, 401–416. [Google Scholar] [CrossRef] [PubMed]
- Shojaei Saadi, H.A.; Gagne, D.; Fournier, E.; Baldoceda Baldeon, L.M.; Sirard, M.A.; Robert, C. Responses of Bovine Early Embryos to S-Adenosyl Methionine Supplementation in Culture. Epigenomics 2016, 8, 1039–1060. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.N.; Hollis, F.; Duclot, F.; Dossat, A.M.; Strong, C.E.; Francis, T.C.; Mercer, R.; Feng, J.; Dietz, D.M.; Lobo, M.K.; et al. Methyl Supplementation Attenuates Cocaine-Seeking Behaviors and Cocaine-Induced C-Fos Activation in a DNA Methylation-Dependent Manner. J. Neurosci. 2015, 35, 8948–8958. [Google Scholar] [CrossRef] [PubMed]
- Maddineni, S.; Nichenametla, S.; Sinha, R.; Wilson, R.P.; Richie, J.P., Jr. Methionine Restriction Affects Oxidative Stress and Glutathione-Related Redox Pathways in the Rat. Exp. Biol. Med. 2013, 238, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yu, L.; Fang, J.; Hu, C.A.; Yin, J.; Ni, H.; Ren, W.; Duraipandiyan, V.; Chen, S.; Al-Dhabi, N.A.; et al. Methionine Restriction on Oxidative Stress and Immune Response in Dss-Induced Colitis Mice. Oncotarget 2017, 8, 44511–44520. [Google Scholar] [CrossRef] [PubMed]
- Ying, Y.; Yun, J.; Guoyao, W.; Kaiji, S.; Zhaolai, D.; Zhenlong, W. Dietary L-Methionine Restriction Decreases Oxidative Stress in Porcine Liver Mitochondria. Exp. Gerontol. 2015, 65, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Roman, I.; Gomez, A.; Perez, I.; Sanchez, C.; Suarez, H.; Naudi, A.; Jove, M.; Lopez-Torres, M.; Pamplona, R.; Barja, G. Effects of Aging and Methionine Restriction Applied at Old Age on Ros Generation and Oxidative Damage in Rat Liver Mitochondria. Biogerontology 2012, 13, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Roman, I.; Gomez, A.; Gomez, J.; Suarez, H.; Sanchez, C.; Naudi, A.; Ayala, V.; Portero-Otin, M.; Lopez-Torres, M.; Pamplona, R.; et al. Forty Percent Methionine Restriction Lowers DNA Methylation, Complex, I. Ros Generation, and Oxidative Damage to Mtdna and Mitochondrial Proteins in Rat Heart. J. Bioenerg. Biomembr. 2011, 43, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Pogribny, I.P.; Basnakian, A.G.; Miller, B.J.; Lopatina, N.G.; Poirier, L.A.; James, S.J. Breaks in Genomic DNA and within the P53 Gene Are Associated with Hypomethylation in Livers of Folate/Methyl-Deficient Rats. Cancer Res. 1995, 55, 1894–1901. [Google Scholar] [PubMed]
- Mattocks, D.A.; Mentch, S.J.; Shneyder, J.; Ables, G.P.; Sun, D.; Richie, J.P., Jr.; Locasale, J.W.; Nichenametla, S.N. Short Term Methionine Restriction Increases Hepatic Global DNA Methylation in Adult but Not Young Male C57bl/6j Mice. Exp. Gerontol. 2017, 88, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nicken, P.; Empl, M.T.; Gerhard, D.; Hausmann, J.; Steinberg, P. Methionine Restriction Inhibits Chemically-Induced Malignant Transformation in the Balb/C 3t3 Cell Transformation Assay. Food Chem. Toxicol. 2016, 95, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E.; Johnson, F.B. Methionine Restriction Activates the Retrograde Response and Confers Both Stress Tolerance and Lifespan Extension to Yeast, Mouse and Human Cells. PLoS ONE 2014, 9, e97729. [Google Scholar] [CrossRef] [PubMed]
- Orgeron, M.L.; Stone, K.P.; Wanders, D.; Cortez, C.C.; Van, N.T.; Gettys, T.W. The Impact of Dietary Methionine Restriction on Biomarkers of Metabolic Health. Prog. Mol. Biol Transl. Sci 2014, 121, 351–376. [Google Scholar] [PubMed] [Green Version]
- Orentreich, N.; Matias, J.R.; DeFelice, A.; Zimmerman, J.A. Low Methionine Ingestion by Rats Extends Life Span. J. Nutr. 1993, 123, 269–274. [Google Scholar] [PubMed]
- James, S.J.; Melnyk, S.; Pogribna, M.; Pogribny, I.P.; Caudill, M.A. Elevation in S-Adenosylhomocysteine and DNA Hypomethylation: Potential Epigenetic Mechanism for Homocysteine-Related Pathology. J. Nutr. 2002, 132, 2361S–2366S. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, D.R.; Marion, D.W.; Cornatzer, W.E.; Duerre, J.A. S-Adenosylmethionine and S-Adenosylhomocystein Metabolism in Isolated Rat Liver. Effects of L.-Methionine, L.-Homocystein, and Adenosine. J. Biol. Chem. 1980, 255, 10822–10827. [Google Scholar] [PubMed]
- Adaikalakoteswari, A.; Finer, S.; Voyias, P.D.; McCarthy, C.M.; Vatish, M.; Moore, J.; Smart-Halajko, M.; Bawazeer, N.; Al-Daghri, N.M.; McTernan, P.G.; et al. Vitamin B12 Insufficiency Induces Cholesterol Biosynthesis by Limiting S-Adenosylmethionine and Modulating the Methylation of Srebf1 and Ldlr Genes. Clin. Epigenet. 2015, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.Y.; Lu, S.C.; Lee, C.M.; Chen, Y.J.; Dugan, T.A.; Huang, W.H.; Chang, S.F.; Liao, W.S.; Chen, C.H.; Lee, Y.T. Homocysteine Inhibits Arterial Endothelial Cell Growth through Transcriptional Downregulation of Fibroblast Growth Factor-2 Involving G Protein and DNA Methylation. Circ. Res. 2008, 102, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Van Hecke, M.V.; Dekker, J.M.; Nijpels, G.; Teerlink, T.; Jakobs, C.; Stolk, R.P.; Heine, R.J.; Bouter, L.M.; Polak, B.C.; Stehouwer, C.D. Homocysteine, S-Adenosylmethionine and S-Adenosylhomocysteine Are Associated with Retinal Microvascular Abnormalities: The Hoorn Study. Clin. Sci. 2008, 114, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, D.; Shima, K.; Matsuo, K.; Nishioka, T.; Chen, C.Y.; Hu, G.F.; Sasaki, A.; Tsuji, T. Ornithine Decarboxylase Antizyme Induces Hypomethylation of Genome DNA and Histone H3 Lysine 9 Dimethylation (H3k9me2) in Human Oral Cancer Cell Line. PLoS ONE 2010, 5, e12554. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, T.; Usui, S.; Aida, T.; Tachikawa, T.; Hu, G.F.; Sasaki, A.; Matsumura, T.; Todd, R.; Wong, D.T. Induction of Epithelial Differentiation and DNA Demethylation in Hamster Malignant Oral Keratinocyte by Ornithine Decarboxylase Antizyme. Oncogene 2001, 20, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E.; Wang, X.; Schwartz, C.E.; McCloskey, D.E. Spermine Synthase Activity Affects the Content of Decarboxylated S-Adenosylmethionine. Biochem. J. 2011, 433, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Shantz, L.M.; Holm, I.; Janne, O.A.; Pegg, A.E. Regulation of S-Adenosylmethionine Decarboxylase Activity by Alterations in the Intracellular Polyamine Content. Biochem. J. 1992, 288 Pt. 2, 511–518. [Google Scholar] [CrossRef]
- Papazafiri, P.; Osborne, H.B. Effect of α-Difluoromethylornithine on DNA Methylation in Murine Erythroleukaemic Cells. Relationship to Stimulation of Induced Differentiation. Biochem. J. 1987, 242, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Poomipark, N.; Flatley, J.E.; Hill, M.H.; Mangnall, B.; Azar, E.; Grabowski, P.; Powers, H.J. Methyl Donor Status Influences Dnmt Expression and Global DNA Methylation in Cervical Cancer Cells. Asian Pac. J. Cancer Prev. 2016, 17, 3213–3222. [Google Scholar] [PubMed]
- Minois, N.; Carmona-Gutierrez, D.; Madeo, F. Polyamines in Aging and Disease. Aging (Albany NY) 2011, 3, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Casillas, M.A., Jr.; Lopatina, N.; Andrews, L.G.; Tollefsbol, T.O. Transcriptional Control of the DNA Methyltransferases Is Altered in Aging and Neoplastically-Transformed Human Fibroblasts. Mol. Cell Biochem. 2003, 252, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Lopatina, N.; Haskell, J.F.; Andrews, L.G.; Poole, J.C.; Saldanha, S.; Tollefsbol, T. Differential Maintenance and De Novo Methylating Activity by Three DNA Methyltransferases in Aging and Immortalized Fibroblasts. J. Cell Biochem. 2002, 84, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Kaplan, M.; Ray, D.; Ray, D.; Zacharek, S.; Gutsch, D.; Richardson, B. Demethylation of ITGAL (Cd11a) Regulatory Sequences in Systemic Lupus Erythematosus. Arthritis Rheum. 2002, 46, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Perez, R.F.; Tejedor, J.R.; Bayon, G.F.; Fernandez, A.F.; Fraga, M.F. Distinct Chromatin Signatures of DNA Hypomethylation in Aging and Cancer. Aging Cell 2018, 17, e12744. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Chen, X.; Ning, C.; Zhu, Q.; Yao, Y.; Zhao, Y.; Luan, F. Methylation of the Genes Rod1, Nlrc5, and Hkr1 Is Associated with Aging in Hainan Centenarians. BMC Med. Genom. 2018, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Florath, I.; Butterbach, K.; Muller, H.; Bewerunge-Hudler, M.; Brenner, H. Cross-Sectional and Longitudinal Changes in DNA Methylation with Age: An Epigenome-Wide Analysis Revealing over 60 Novel Age-Associated Cpg Sites. Hum. Mol. Genet. 2014, 23, 1186–1201. [Google Scholar] [CrossRef] [PubMed]
- McClay, J.L.; Aberg, K.A.; Clark, S.L.; Nerella, S.; Kumar, G.; Xie, L.Y.; Hudson, A.D.; Harada, A.; Hultman, C.M.; Magnusson, P.K.; et al. A Methylome-Wide Study of Aging Using Massively Parallel Sequencing of the Methyl-Cpg-Enriched Genomic Fraction from Blood in over 700 Subjects. Hum. Mol. Genet. 2014, 23, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Alisch, R.S.; Barwick, B.G.; Chopra, P.; Myrick, L.K.; Satten, G.A.; Conneely, K.N.; Warren, S.T. Age-Associated DNA Methylation in Pediatric Populations. Genome Res. 2012, 22, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Richardson, B.C. Role of DNA Methylation in the Regulation of Cell Function: Autoimmunity, Aging and Cancer. J. Nutr. 2002, 132, 2401S–2405S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatazawa, Y.; Ono, Y.; Hirose, Y.; Kanai, S.; Fujii, N.L.; Machida, S.; Nishino, I.; Shimizu, T.; Okano, M.; Kamei, Y.; et al. Reduced Dnmt3a Increases Gdf5 Expression with Suppressed Satellite Cell Differentiation and Impaired Skeletal Muscle Regeneration. FASEB J. 2018, 32, 1452–1467. [Google Scholar] [CrossRef] [PubMed]
- De Magalhaes, J.P. How Ageing Processes Influence Cancer. Nat. Rev. Cancer 2013, 13, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.C.; Houseman, E.A.; King, J.E.; Christensen, B.C. Normal Breast Tissue DNA Methylation Differences at Regulatory Elements Are Associated with the Cancer Risk Factor Age. Breast Cancer Res. 2017, 19, 81. [Google Scholar] [CrossRef] [PubMed]
- Galamb, O.; Kalmár, A.; Barták, B.K.; Patai, Á.V.; Leiszter, K.; Péterfia, B.; Wichmann, B.; Valcz, G.; Veres, G.; Tulassay, Z.; et al. Aging Related Methylation Influences the Gene Expression of Key Control Genes in Colorectal Cancer and Adenoma. World J. Gastroenterol. 2016, 22, 10325–10340. [Google Scholar] [CrossRef] [PubMed]
- Okuchi, Y.; Imajo, M.; Mizuno, R.; Kamioka, Y.; Miyoshi, H.; Taketo, M.M.; Nagayama, S.; Sakai, Y.; Matsuda, M. Identification of Aging-Associated Gene Expression Signatures That Precede Intestinal Tumorigenesis. PLoS ONE 2016, 11, e0162300. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wagner, W. Epigenetic Aging Signatures Are Coherently Modified in Cancer. PLoS Genet. 2015, 11, e1005334. [Google Scholar] [CrossRef] [PubMed]
- Wada, H.; Wada, U.F.; Mano, H.; Higashiguchi, M.; Haba, R.; Watanabe, S.; Udaka, S. Effects of Dietary Polyamines on the Promotion of Mammary Tumor in Rats. J. Health Sci. 2002, 48, 376–380. [Google Scholar] [CrossRef] [Green Version]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of C-Myc as a Target of the Apc Pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Bello-Fernandez, C.; Packham, G.; Cleveland, J.L. The Ornithine Decarboxylase Gene Is a Transcriptional Target of C-Myc. Proc. Natl. Acad. Sci. USA 1993, 90, 7804–7808. [Google Scholar] [CrossRef] [PubMed]
- Pena, A.; Reddy, C.D.; Wu, S.; Hickok, N.J.; Reddy, E.P.; Yumet, G.; Soprano, D.R.; Soprano, K.J. Regulation of Human Ornithine Decarboxylase Expression by the C-Myc.Max Protein Complex. J. Biol. Chem. 1993, 268, 27277–27285. [Google Scholar] [PubMed]
- Shukla-Dave, A.; Castillo-Martin, M.; Chen, M.; Lobo, J.; Gladoun, N.; Collazo-Lorduy, A.; Khan, F.M.; Ponomarev, V.; Yi, Z.; Zhang, W.; et al. Ornithine Decarboxylase Is Sufficient for Prostate Tumorigenesis via Androgen Receptor Signaling. Am. J. Pathol. 2016, 186, 3131–3145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auvinen, M.; Paasinen, A.; Andersson, L.C.; Holtta, E. Ornithine Decarboxylase Activity Is Critical for Cell Transformation. Nature 1992, 360, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Erdman, S.H.; Ignatenko, N.A.; Powell, M.B.; Blohm-Mangone, K.A.; Holubec, H.; Guillen-Rodriguez, J.M.; Gerner, E.W. Apc-Dependent Changes in Expression of Genes Influencing Polyamine Metabolism, and Consequences for Gastrointestinal Carcinogenesis, in the Min Mouse. Carcinogenesis 1999, 20, 1709–1713. [Google Scholar] [CrossRef] [PubMed]
- Megosh, L.; Halpern, M.; Farkash, E.; O’Brien, T.G. Analysis of Ras Gene Mutational Spectra in Epidermal Papillomas from K6/Odc Transgenic Mice. Mol. Carcinog. 1998, 22, 145–149. [Google Scholar] [CrossRef]
- Moshier, J.A.; Malecka-Panas, E.; Geng, H.; Dosescu, J.; Tureaud, J.; Skunca, M.; Majumdar, A.P. Ornithine Decarboxylase Transformation of NIH/3T3 Cells Is Mediated by Altered Epidermal Growth Factor Receptor Activity. Cancer Res. 1995, 55, 5358–5365. [Google Scholar] [PubMed]
- Shantz, L.M.; Pegg, A.E. Ornithine Decarboxylase Induction in Transformation by H-Ras and Rhoa. Cancer Res. 1998, 58, 2748–2753. [Google Scholar] [PubMed]
- Holtta, E.; Sistonen, L.; Alitalo, K. The Mechanisms of Ornithine Decarboxylase Deregulation in C-Ha-Ras Oncogene-Transformed Nih 3t3 Cells. J. Biol. Chem. 1988, 263, 4500–4507. [Google Scholar] [PubMed]
- Clifford, A.; Morgan, D.; Yuspa, S.H.; Soler, A.P.; Gilmour, S. Role of Ornithine Decarboxylase in Epidermal Tumorigenesis. Cancer Res. 1995, 55, 1680–1686. [Google Scholar] [PubMed]
- Hibshoosh, H.; Johnson, M.; Weinstein, I.B. Effects of Overexpression of Ornithine Decarboxylase (Odc) on Growth Control and Oncogene-Induced Cell Transformation. Oncogene 1991, 6, 739–743. [Google Scholar]
- O’Brien, T.G.; Megosh, L.C.; Gilliard, G.; Soler, A.P. Ornithine Decarboxylase Overexpression Is a Sufficient Condition for Tumor Promotion in Mouse Skin. Cancer Res. 1997, 57, 2630–2637. [Google Scholar] [PubMed]
- Ito, K.; Kashiwagi, K.; Watanabe, S.; Kameji, T.; Hayashi, S.; Igarashi, K. Influence of the 5′-Untranslated Region of Ornithine Decarboxylase Mrna and Spermidine on Ornithine Decarboxylase Synthesis. J. Biol. Chem. 1990, 265, 13036–13041. [Google Scholar] [PubMed]
- Kashiwagi, K.; Ito, K.; Igarashi, K. Spermidine Regulation of Ornithine Decarboxylase Synthesis by a Gc-Rich Sequence of the 5′-Untranslated Region. Biochem. Biophys. Res. Commun. 1991, 178, 815–822. [Google Scholar] [CrossRef]
- Lovkvist, E.; Stjernborg, L.; Persson, L. Feedback Regulation of Mammalian Ornithine Decarboxylase. Studies Using a Transient Expression System. Eur J. Biochem. 1993, 215, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Vargas, A.J.; Wertheim, B.C.; Gerner, E.W.; Thomson, C.A.; Rock, C.L.; Thompson, P.A. Dietary Polyamine Intake and Risk of Colorectal Adenomatous Polyps. Am. J. Clin. Nutr. 2012, 96, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Vargas, A.J.; Ashbeck, E.L.; Wertheim, B.C.; Wallace, R.B.; Neuhouser, M.L.; Thomson, C.A.; Thompson, P.A. Dietary Polyamine Intake and Colorectal Cancer Risk in Postmenopausal Women. Am. J. Clin. Nutr. 2015, 102, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Huidobro, C.; Fernandez, A.F.; Fraga, M.F. Aging Epigenetics: Causes and Consequences. Mol. Aspects Med. 2013, 34, 765–781. [Google Scholar] [CrossRef] [PubMed]
- Maeda, M.; Nakajima, T.; Oda, I.; Shimazu, T.; Yamamichi, N.; Maekita, T.; Asada, K.; Yokoi, C.; Ando, T.; Yoshida, T.; et al. High Impact of Methylation Accumulation on Metachronous Gastric Cancer: 5-Year Follow-up of a Multicentre Prospective Cohort Study. Gut 2017, 66, 1721–1723. [Google Scholar] [CrossRef] [PubMed]
- Hai, Z.; Zuo, W. Aberrant DNA Methylation in the Pathogenesis of Atherosclerosis. Clin. Chim. Acta 2016, 456, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Maegawa, S.; Gough, S.M.; Watanabe-Okochi, N.; Lu, Y.; Zhang, N.; Castoro, R.J.; Estecio, M.R.; Jelinek, J.; Liang, S.; Kitamura, T.; et al. Age-Related Epigenetic Drift in the Pathogenesis of Mds and Aml. Genome Res. 2014, 24, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Borghini, A.; Cervelli, T.; Galli, A.; Andreassi, M.G. DNA Modifications in Atherosclerosis: From the Past to the Future. Atherosclerosis 2013, 230, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Vlaming, H.; van Leeuwen, F. Crosstalk between Aging and the Epigenome. Epigenomics 2012, 4, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Nojima, M.; Iwasaki, M.; Kasuga, Y.; Yokoyama, S.; Onuma, H.; Nishimura, H.; Kusama, R.; Yoshida, T.; Tsugane, S. Correlation between Global Methylation Level of Peripheral Blood Leukocytes and Serum C Reactive Protein Level Modified by Mthfr Polymorphism: A Cross-Sectional Study. BMC Cancer 2018, 18, 184. [Google Scholar] [CrossRef] [PubMed]
- Iwaya, C.; Kitajima, H.; Yamamoto, K.; Maeda, Y.; Sonoda, N.; Shibata, H.; Inoguchi, T. DNA Methylation of the Klf14 Gene Region in Whole Blood Cells Provides Prediction for the Chronic Inflammation in the Adipose Tissue. Biochem. Biophys. Res. Commun. 2018, 497, 908–915. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.; Zhang, Q.H.; Tang, Q.; Jiang, Z.; Xing, S.; Li, J.; Pang, Y. Comprehensive Analysis of Gene Expression and DNA Methylation Datasets Identify Valuable Biomarkers for Rheumatoid Arthritis Progression. Oncotarget 2018, 9, 2977–2983. [Google Scholar] [CrossRef] [PubMed]
- Kan, S.; Wu, J.; Sun, C.; Hao, J.; Wu, Z. Correlation between Rage Gene Promoter Methylation and Diabetic Retinal Inflammation. Exp. Ther. Med. 2018, 15, 242–246. [Google Scholar] [CrossRef] [PubMed]




| Activity | Authors | Journal (Year) |
|---|---|---|
| Anti-inflammation | Lovaas E. et al. | Free. Radic. Biol. Med. 1991, 11, 455–461. [111] |
| Zhang M. et al. | J. Exp. Med. 1997, 185, 1759–1768. [74] | |
| Soda K. et al. | J. Immunol. 2005, 175, 237–245. [19] | |
| Lagishetty C.V. et al. | Indian. J. Pharmacol. 2008, 40, 121–125. [112] | |
| Choi Y.H. et al. | J. Biomed. Sci. 2012, 19, 31. [113] | |
| Paul S. et al. | Inflamm. Res. 2013, 62, 681–688. [114] | |
| Zhou S. et al. | Front. Immunol. 2018, 9, 948. [115] | |
| Anti-oxidant & Free radical scavenger | Tadolini B. et al. | Biochem. Biophys. Res. Commun. 1984, 122, 550–555. [116] |
| Lovaas E. et al. | Free Radic. Biol. Med. 1991, 11, 455–461. [111] | |
| Khan A.U. et al. | Proc. Natl. Acad. Sci. USA 1992, 89, 11428–11430. [117] | |
| Goss S.P. et al. | Chem. Res. Toxicol. 1995, 8, 800–806. [118] | |
| Marzabadi M.R. et al. | Free. Radic. Biol. Med. 1996, 21, 375–381. [119] | |
| Farbiszewski R. et al. | Neurochem. Res. 1996, 21, 1497–1503. [120] | |
| Ha H.C. et al. | Proc. Natl. Acad. Sci. USA 1998, 95, 11140–11145. [121] | |
| Jung I.L. et al. | Arch. Biochem. Biophys. 2003, 418, 125–132. [122] | |
| Chattopadhyay M.K. et al. | Proc. Natl. Acad. Sci. USA 2003, 100, 2261–2265. [123] | |
| Belle N.A. et al. | Brain Res. 2004, 1008, 245–251. [124] | |
| Gaboriau F. et al. | Redox. Rep. 2005, 10, 9–18. [125] | |
| Fujisawa S. et al. | Anticancer Res. 2005, 25, 965–969. [126] | |
| Sava I.G. et al. | Free Radic. Biol. Med. 2006, 41, 1272–1281. [127] | |
| Rider J.E. et al. | Amino. Acids 2007, 33, 231–240. [128] | |
| Nayvelt I. et al. | Biomacromolecules 2010, 11, 97–105. [129] | |
| Jeong J.W. et al. | Biomol. Ther. (Seoul) 2018, 26, 146–156. [130] | |
| Radioprotection | Courdi A. et al. | Int. J. Cancer 1986, 38, 103–107. [131] |
| Arundel C.M. et al. | Radiat. Res. 1988, 114, 634–640. [132] | |
| Held K.D. et al. | Int. J. Radiat. Biol. 1991, 59, 699–710. [133] | |
| Snyder R.D. et al. | Radiat. Res. 1994, 137, 67–75. [134] | |
| Williams J.R. et al. | Biochem. Biophys. Res. Commun. 1994, 201, 1–7. [135] | |
| Spotheim-Maurizot M. et al. | Int. J. Radiat. Biol. 1995, 68, 571–577. [136] | |
| Newton G.L. et al. | Radiat. Res. 1996, 145, 776–780. [137] | |
| Chiu S. et al. | Radiat. Res. 1998, 149, 543–549. [138] | |
| Sy D. et al. | Int. J. Radiat. Biol. 1999, 75, 953–961. [139] | |
| Warters R.L. et al. | Radiat. Res. 1999, 151, 354–362. [140] | |
| Douki T. et al. | Radiat. Res. 2000, 153, 29–35. [141] | |
| von Deutsch A.W. et al. | Gravit. Space Biol. Bull. 2005, 18, 109–110. [142] | |
| Protection from ultraviolet light | Snyder R.D. et al. | Photochem. Photobiol. 1990, 52, 525–532. [143] |
| Williams J.R. et al. | Biochem. Biophys. Res. Commun. 1994, 201, 1–7. [144] | |
| Pothipongsa A. et al. | Appl. Biochem. Biotechnol. 2012, 168, 1476–1488. [145] | |
| Protection from chemicals & other stress | Rajalakshmi S. et al. | Biochemistry. 1978, 17, 4515–4518. [144] |
| Mackintosh C.A. et al. | Biochem. J. 2000, 351, 439–447. [146] | |
| Di Mascio P. et al. | Arch. Biochem. Biophys. 2000, 373, 368–374. [147] | |
| Chauhan S.D. et al. | FASEB. J. 2003, 17, 773–775. [148] | |
| Gugliucci A. et al. | Life Sci. 2003, 72, 2603–2616. [149] | |
| Sagor G.H. et al. | Transgenic Res. 2013, 22, 595–605. [150] | |
| Okumura S. et al. | Liver Transpl. 2016, 22, 1231–1244. [151] |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soda, K. Polyamine Metabolism and Gene Methylation in Conjunction with One-Carbon Metabolism. Int. J. Mol. Sci. 2018, 19, 3106. https://doi.org/10.3390/ijms19103106
Soda K. Polyamine Metabolism and Gene Methylation in Conjunction with One-Carbon Metabolism. International Journal of Molecular Sciences. 2018; 19(10):3106. https://doi.org/10.3390/ijms19103106
Chicago/Turabian StyleSoda, Kuniyasu. 2018. "Polyamine Metabolism and Gene Methylation in Conjunction with One-Carbon Metabolism" International Journal of Molecular Sciences 19, no. 10: 3106. https://doi.org/10.3390/ijms19103106
APA StyleSoda, K. (2018). Polyamine Metabolism and Gene Methylation in Conjunction with One-Carbon Metabolism. International Journal of Molecular Sciences, 19(10), 3106. https://doi.org/10.3390/ijms19103106