Generation of a 3D Liver Model Comprising Human Extracellular Matrix in an Alginate/Gelatin-Based Bioink by Extrusion Bioprinting for Infection and Transduction Studies
Abstract
:1. Introduction
2. Results and Discussion
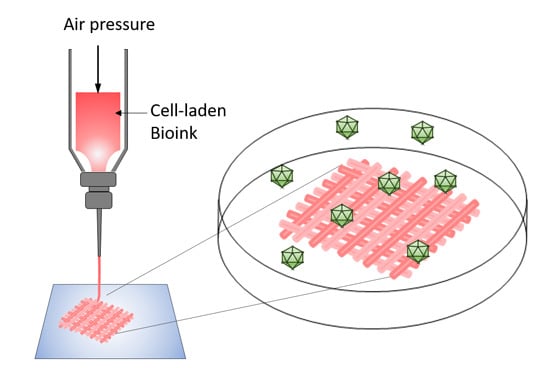
2.1. Preparation and Characterization of HepaRG Cell-Laden Hydrogels Containing Varying Amounts of Extracellular Matrix (ECM)
2.2. Characterization of Hepatic Metabolism in HepaRG Cell-Laden Bioinks Containing Various Amounts of ECM
2.3. Characterization of Rheological Properties of ECM-Based Hydrogels
2.4. Transduction of Bioprinted Liver Model with Adeno-Associated Virus (AAV) Vectors
2.5. Adenovirus Infection of Bioprinted Liver Model
3. Materials and Methods
3.1. Cell Culture and Human Extracellular Matrix Preparation
3.2. Preparation of Cell-Laden Biopolymers
3.3. 3D Bioprinting
3.4. Rheological Properties
3.5. Cell Distribution
3.6. Cell Viability and Lactate Dehydrogenase Release
3.7. Albumin Secretion and Cytochrome P450 3A4 Activity
3.8. Adeno-Associated Virus Production, Transduction, and hCycB Knockdown
3.9. Human Adenovirus 5 Preparation and Infection
3.10. Cell-Killing Assay
3.11. Reverse Transcription and Quantitative Polymerase Chain Reaction (qPCR)
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AAV | Adeno-associated virus |
| AAV2.6 | Pseudotyped scAAV vector of serotype 6 |
| AdV | Adenovirus |
| hAdV5 | Human adenovirus serotype 5 |
| MOI | Multiplicity of infection |
| ECM | Extracellular matrix |
| hECM | Human extracellular matrix |
| hCycB | Human cyclophillin b |
| qPCR | Quantitative polymerase chain reaction |
| RNAi | RNA interferencelinear dichroism |
| shRNA | Small hairpin RNA |
| EmGFP | Emerald green fluorescent protein |
| CYP3A4 | Cytochrome P450 oxidase 3A4 |
References
- Guha, C.; Mohan, S.; Roy-Chowdhury, N.; Roy-Chowdhury, J. Cell culture and animal models of viral hepatitis. Part I: Hepatitis B. Lab. Anim. (NY) 2004, 33, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Hough, R.; Chetwood, A.; Sinfield, R.; Welch, J.; Vora, A. Fatal adenovirus hepatitis during standard chemotherapy for childhood acute lymphoblastic leukemia. J. Pediatr. Hematol. Oncol. 2005, 27, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, N.M.; Lowen, A.C. Animal Models for Influenza Virus Pathogenesis and Transmission. Viruses 2010, 2, 1530–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaar, K.; Geisler, A.; Kraus, M.; Pinkert, S.; Pryshliak, M.; Spencer, J.F.; Tollefson, A.E.; Ying, B.; Kurreck, J.; Wold, W.S.; et al. Anti-adenoviral Artificial MicroRNAs Expressed from AAV9 Vectors Inhibit Human Adenovirus Infection in Immunosuppressed Syrian Hamsters. Mol. Ther. Nucleic Acids 2017, 8, 300–316. [Google Scholar] [CrossRef] [PubMed]
- Berk, A.J. Adenoviridae: The viruses and Their Replication. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippinocott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 2355–2394. [Google Scholar]
- Baldwin, A.; Kingman, H.; Darville, M.; Foot, A.B.; Grier, D.; Cornish, J.M.; Goulden, N.; Oakhill, A.; Pamphilon, D.H.; Steward, C.G.; et al. Outcome and clinical course of 100 patients with adenovirus infection following bone marrow transplantation. Bone Marrow Transplant. 2000, 26, 1333–1338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kay, M.A. State-of-the-art gene-based therapies: The road ahead. Nat. Rev. Genet. 2011, 12, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Voretigene neparvovec-rzyl (Luxturna) for inherited retinal dystrophy. Med. Lett. Drugs Ther. 2018, 60, 53–55.
- Yla-Herttuala, S. Endgame: Glybera finally recommended for approval as the first gene therapy drug in the European union. Mol. Ther. 2010, 20, 1831–1832. [Google Scholar] [CrossRef] [PubMed]
- Lisowski, L.; Tay, S.S.; Alexander, I.E. Adeno-associated virus serotypes for gene therapeutics. Curr. Opin. Pharmacol. 2015, 24, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Enger, P.O.; Thorsen, F.; Lonning, P.E.; Bjerkvig, R.; Hoover, F. Adeno-associated viral vectors penetrate human solid tumor tissue in vivo more effectively than adenoviral vectors. Hum. Gene Ther. 2002, 13, 1115–1125. [Google Scholar] [CrossRef] [PubMed]
- Thorsen, F.; Afione, S.; Huszthy, P.C.; Tysnes, B.B.; Svendsen, A.; Bjerkvig, R.; Kotin, R.M.; Lonning, P.E.; Hoover, F. Adeno-associated virus (AAV) serotypes 2, 4 and 5 display similar transduction profiles and penetrate solid tumor tissue in models of human glioma. J. Gene Med. 2006, 8, 1131–1140. [Google Scholar] [CrossRef] [PubMed]
- Grix, T.; Ruppelt, A.; Thomas, A.; Amler, A.K.; Noichl, B.P.; Lauster, R.; Kloke, L. Bioprinting Perfusion-Enabled Liver Equivalents for Advanced Organ-on-a-Chip Applications. Genes (Basel) 2018, 9, 176. [Google Scholar] [CrossRef] [PubMed]
- Ozbolat, I.T.; Peng, W.; Ozbolat, V. Application areas of 3D bioprinting. Drug Discov. Today 2016, 21, 1257–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, F.; Eames, B.F.; Chen, X. Application of Extrusion-Based Hydrogel Bioprinting for Cartilage Tissue Engineering. Int. J. Mol. Sci. 2017, 18, E1597. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yan, Y.; Pan, Y.; Xiong, Z.; Liu, H.; Cheng, J.; Liu, F.; Lin, F.; Wu, R.; Zhang, R.; et al. Generation of three-dimensional hepatocyte/gelatin structures with rapid prototyping system. Tissue Eng. 2006, 12, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, H. Incorporation of DMSO and dextran-40 into a gelatin/alginate hydrogel for controlled assembled cell cryopreservation. Cryobiology 2010, 61, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Billiet, T.; Gevaert, E.; De Schryver, T.; Cornelissen, M.; Dubruel, P. The 3D printing of gelatin methacrylamide cell-laden tissue-engineered constructs with high cell viability. Biomaterials 2014, 35, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Axpe, E.; Oyen, M.L. Applications of Alginate-Based Bioinks in 3D Bioprinting. Int. J. Mol. Sci. 2016, 17, E1976. [Google Scholar] [CrossRef] [PubMed]
- Hospodiuk, M.; Dey, M.; Sosnoski, D.; Ozbolat, I.T. The bioink: A comprehensive review on bioprintable materials. Biotechnol. Adv. 2017, 35, 217–239. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, D.; Tun, H.W.; Wang, T.; Naing, M.W. Organ-Derived Decellularized Extracellular Matrix: A Game Changer for Bioink Manufacturing? Trends Biotechnol. 2018, 36, 787–805. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Han, W.; Kim, H.; Ha, D.H.; Jang, J.; Kim, B.S.; Cho, D.W. Development of Liver Decellularized Extracellular Matrix Bioink for Three-Dimensional Cell Printing-Based Liver Tissue Engineering. Biomacromolecules 2017, 18, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Kleinman, H.K.; McGarvey, M.L.; Liotta, L.A.; Robey, P.G.; Tryggvason, K.; Martin, G.R. Isolation and characterization of type IV procollagen, laminin, and heparan sulfate proteoglycan from the EHS sarcoma. Biochemistry 1982, 21, 6188–6193. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.S.; Postovit, L.M.; Lajoie, G.A. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics 2010, 10, 1886–1890. [Google Scholar] [CrossRef] [PubMed]
- Kleinman, H.K.; Martin, G.R. Matrigel: Basement membrane matrix with biological activity. Semin. Cancer Biol. 2005, 15, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Benton, G.; Arnaoutova, I.; George, J.; Kleinman, H.K.; Koblinski, J. Matrigel: From discovery and ECM mimicry to assays and models for cancer research. Adv. Drug Deliv. Rev. 2014, 79–80, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Eo, J.S.; Soo, H.J.; Jong-Ock, S.; Kyung-Hwan, J.; Nam-Soo, K. Extracellular Matrix and 3D Printing. Curr. Trends Biomed. Eng. Biosci. 2017, 2, 55596. [Google Scholar]
- Nibourg, G.A.; Chamuleau, R.A.; van Gulik, T.M.; Hoekstra, R. Proliferative human cell sources applied as biocomponent in bioartificial livers: A review. Expert Opin. Biol. Ther. 2012, 12, 905–921. [Google Scholar] [CrossRef] [PubMed]
- Aninat, C.; Piton, A.; Glaise, D.; Le Charpentier, T.; Langouet, S.; Morel, F.; Guguen-Guillouzo, C.; Guillouzo, A. Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab. Dispos. 2006, 34, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Kanebratt, K.P.; Andersson, T.B. Evaluation of HepaRG cells as an in vitro model for human drug metabolism studies. Drug Metab. Dispos. 2008, 36, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Marion, M.J.; Hantz, O.; Durantel, D. The HepaRG cell line: Biological properties and relevance as a tool for cell biology, drug metabolism, and virology studies. Methods Mol. Biol. 2010, 640, 261–272. [Google Scholar] [PubMed]
- Rahali, K.; Ben Messaoud, G.; Kahn, C.J.F.; Sanchez-Gonzalez, L.; Kaci, M.; Cleymand, F.; Fleutot, S.; Linder, M.; Desobry, S.; Arab-Tehrany, E. Synthesis and Characterization of Nanofunctionalized Gelatin Methacrylate Hydrogels. Int. J. Mol. Sci. 2017, 18, 2675. [Google Scholar] [CrossRef] [PubMed]
- You, F.; Wu, X.; Chen, D.X.B. 3D printing of porous alginate/gelatin hydrogel scaffolds and their mechanical property characterization. Int. J. Pol. Mater. Pol. Biomater. 2016, 66, 299–306. [Google Scholar] [CrossRef]
- Lou, Y.; Li, Y.; Qin, X.; Wa, Q. 3D printing of concentrated alginate/gelatin scaffolds with homogeneous nano apatite coating for bone tissue engineering. Mater. Des. 2018, 146, 12–19. [Google Scholar]
- Berg, J.; Hiller, T.; Kissner, M.S.; Qazi, T.H.; Duda, G.N.; Hocke, A.C.; Hippenstiel, S.; Elomaa, L.; Weinhart, M.; Fahrenson, C.; et al. Optimization of cell-laden bioinks for 3D bioprinting and efficient infection with influenza A virus. Sci. Rep. 2018, 8, 13877. [Google Scholar] [CrossRef] [PubMed]
- Denner, J.; Tonjes, R.R. Infection barriers to successful xenotransplantation focusing on porcine endogenous retroviruses. Clin. Microbiol. Rev. 2012, 25, 318–343. [Google Scholar] [CrossRef] [PubMed]
- Beachley, V.Z.; Wolf, M.T.; Sadtler, K.; Manda, S.S.; Jacobs, H.; Blatchley, M.R.; Bader, J.S.; Pandey, A.; Pardoll, D.; Elisseeff, J.H. Tissue matrix arrays for high-throughput screening and systems analysis of cell function. Nat. Methods 2015, 12, 1197–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gelse, K.; Poschl, E.; Aigner, T. Collagens—Structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003, 55, 1531–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miron-Mendoza, M.; Seemann, J.; Grinnell, F. The differential regulation of cell motile activity through matrix stiffness and porosity in three dimensional collagen matrices. Biomaterials 2010, 31, 6425–6435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antoine, E.E.; Vlachos, P.P.; Rylander, M.N. Review of collagen I hydrogels for bioengineered tissue microenvironments: Characterization of mechanics, structure, and transport. Tissue Eng. Part. B Rev. 2014, 20, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.K.; Hoying, J.B. Bioprinting in Regenrative Medicine; Turkse, K., Ed.; Springer: Cham, Switzerland; Heidelberg, Germany; New York, NY, USA; Dordrecht, The Netherlands; London, UK, 2015; pp. 1–31. [Google Scholar]
- Roth, E.A.; Xu, T.; Das, M.; Gregory, C.; Hickman, J.J.; Boland, T. Inkjet printing for high-throughput cell patterning. Biomaterials 2004, 25, 3707–3715. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.J.; Su, X.; Xu, Y.Y.; Kong, B.; Sun, W.; Mi, S.L. Bioprinting three-dimensional cell-laden tissue constructs with controllable degradation. Sci. Rep. 2016, 6, 24474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, V.; Singh, G.; Trasatti, J.P.; Bjornsson, C.; Xu, X.; Tran, T.N.; Yoo, S.S.; Dai, G.; Karande, P. Design and fabrication of human skin by three-dimensional bioprinting. Tissue Eng. Part. C Methods 2014, 20, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Choi, J.C.; Shim, J.H.; Lee, J.S.; Park, H.; Kim, S.W.; Doh, J.; Cho, D.W. A comparative study on collagen type I and hyaluronic acid dependent cell behavior for osteochondral tissue bioprinting. Biofabrication 2014, 6, 035004. [Google Scholar] [CrossRef] [PubMed]
- Cross, V.L.; Zheng, Y.; Won Choi, N.; Verbridge, S.S.; Sutermaster, B.A.; Bonassar, L.J.; Fischbach, C.; Stroock, A.D. Dense type I collagen matrices that support cellular remodeling and microfabrication for studies of tumor angiogenesis and vasculogenesis in vitro. Biomaterials 2010, 31, 8596–8607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basheer, L.; Kerem, Z. Interactions between CYP3A4 and Dietary Polyphenols. Oxid. Med. Cell. Longev. 2015, 2015, 854015. [Google Scholar] [CrossRef] [PubMed]
- Raoufinia, R.; Mota, A.; Keyhanvar, N.; Safari, F.; Shamekhi, S.; Abdolalizadeh, J. Overview of Albumin and Its Purification Methods. Adv. Pharm. Bull. 2016, 6, 495–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gripon, P.; Rumin, S.; Urban, S.; Le Seyec, J.; Glaise, D.; Cannie, I.; Guyomard, C.; Lucas, J.; Trepo, C.; Guguen-Guillouzo, C. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. USA 2002, 99, 15655–15660. [Google Scholar] [CrossRef] [PubMed]
- Parent, R.; Marion, M.J.; Furio, L.; Trepo, C.; Petit, M.A. Origin and characterization of a human bipotent liver progenitor cell line. Gastroenterology 2004, 126, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, R.; Nibourg, G.A.; van der Hoeven, T.V.; Ackermans, M.T.; Hakvoort, T.B.; van Gulik, T.M.; Lamers, W.H.; Elferink, R.P.; Chamuleau, R.A. The HepaRG cell line is suitable for bioartificial liver application. Int. J. Biochem. Cell. Biol. 2011, 43, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Gasperini, L.; Mano, J.F.; Reis, R.L. Natural polymers for the microencapsulation of cells. J. R. Soc. Interface 2014, 11, 20140817. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.J.; Henstock, P.V.; Dunn, M.C.; Smith, A.R.; Chabot, J.R.; de Graaf, D. Cellular imaging predictions of clinical drug-induced liver injury. Toxicol. Sci. 2008, 105, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, E.; Walsky, R.; Zhang, H.; Bi, Y.A.; Whalen, K.M.; Yang, Y.S.; Linder, C.; Xiao, Y.; Iseki, K.; Fenner, K.S.; et al. Differential modulation of cytochrome P450 activity and the effect of 1-aminobenzotriazole on hepatic transport in sandwich-cultured human hepatocytes. Drug Metab. Dispos. 2012, 40, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Lauschke, V.M.; Hendriks, D.F.; Bell, C.C.; Andersson, T.B.; Ingelman-Sundberg, M. Novel 3D Culture Systems for Studies of Human Liver Function and Assessments of the Hepatotoxicity of Drugs and Drug Candidates. Chem. Res. Toxicol. 2016, 29, 1936–1955. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wei, D.; Yang, K.; Liu, X.; Hongsong, F.; Zhang, X. The development of cell-initiated degradable hydrogel based on methacrylated alginate applicable to multiple microfabrication technologies. J. Mater. Chem. B 2017, 5, 8060–8069. [Google Scholar] [CrossRef]
- Gunness, P.; Mueller, D.; Shevchenko, V.; Heinzle, E.; Ingelman-Sundberg, M.; Noor, F. 3D organotypic cultures of human HepaRG cells: A tool for in vitro toxicity studies. Toxicol. Sci. 2013, 133, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Hori, Y.; Yamamoto, T.; Urashima, T.; Ohara, Y.; Tanaka, H. 3D spheroid cultures improve the metabolic gene expression profiles of HepaRG cells. Biosci. Rep. 2015, 35, e00208. [Google Scholar] [CrossRef] [PubMed]
- Mironov, V.; Visconti, R.P.; Kasyanov, V.; Forgacs, G.; Drake, C.J.; Markwald, R.R. Organ printing: Tissue spheroids as building blocks. Biomaterials 2009, 30, 2164–2174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fechner, H.; Pinkert, S.; Geisler, A.; Poller, W.; Kurreck, J. Pharmacological and biological antiviral therapeutics for cardiac coxsackievirus infections. Molecules 2011, 16, 8475–8503. [Google Scholar] [CrossRef] [PubMed]
- Kay, M.A.; Nakai, H. Looking into the safety of AAV vectors. Nature 2003, 424, 251. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Ortiz, J.L.; Schaffer, D.V. Adeno-associated virus (AAV) vectors in cancer gene therapy. J. Control. Release 2016, 240, 287–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimm, D.; Kay, M.A. From virus evolution to vector revolution: Use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr. Gene Ther. 2003, 3, 281–304. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Kay, M.A. RNAi and Gene Therapy: A Mutual Attraction. Hematol. Am. Soc. Hematol. Educ. Program. 2007, 2007, 473–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Lillicrap, D.; Patarroyo-White, S.; Liu, T.; Qian, X.; Scallan, C.D.; Powell, S.; Keller, T.; McMurray, M.; Labelle, A.; et al. Multiyear therapeutic benefit of AAV serotypes 2, 6, and 8 delivering factor VIII to hemophilia A mice and dogs. Blood 2006, 108, 107–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiller, T.; Rohrs, V.; Dehne, E.M.; Wagner, A.; Fechner, H.; Lauster, R.; Kurreck, J. Study of Viral Vectors in a Three-dimensional Liver Model Repopulated with the Human Hepatocellular Carcinoma Cell Line HepG2. J. Vis. Exp. 2016, 116, e54633. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Rohrs, V.; Materne, E.M.; Hiller, T.; Kedzierski, R.; Fechner, H.; Lauster, R.; Kurreck, J. Use of a three-dimensional humanized liver model for the study of viral gene vectors. J. Biotechnol. 2015, 212, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Rohrs, V.; Kedzierski, R.; Fechner, H.; Kurreck, J. A novel method for the quantification of adeno-associated virus vectors for RNA interference applications using quantitative polymerase chain reaction and purified genomic adeno-associated virus DNA as a standard. Hum. Gene Ther. Methods 2013, 24, 355–363. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hiller, T.; Berg, J.; Elomaa, L.; Röhrs, V.; Ullah, I.; Schaar, K.; Dietrich, A.-C.; Al-Zeer, M.A.; Kurtz, A.; Hocke, A.C.; et al. Generation of a 3D Liver Model Comprising Human Extracellular Matrix in an Alginate/Gelatin-Based Bioink by Extrusion Bioprinting for Infection and Transduction Studies. Int. J. Mol. Sci. 2018, 19, 3129. https://doi.org/10.3390/ijms19103129
Hiller T, Berg J, Elomaa L, Röhrs V, Ullah I, Schaar K, Dietrich A-C, Al-Zeer MA, Kurtz A, Hocke AC, et al. Generation of a 3D Liver Model Comprising Human Extracellular Matrix in an Alginate/Gelatin-Based Bioink by Extrusion Bioprinting for Infection and Transduction Studies. International Journal of Molecular Sciences. 2018; 19(10):3129. https://doi.org/10.3390/ijms19103129
Chicago/Turabian StyleHiller, Thomas, Johanna Berg, Laura Elomaa, Viola Röhrs, Imran Ullah, Katrin Schaar, Ann-Christin Dietrich, Munir A. Al-Zeer, Andreas Kurtz, Andreas C. Hocke, and et al. 2018. "Generation of a 3D Liver Model Comprising Human Extracellular Matrix in an Alginate/Gelatin-Based Bioink by Extrusion Bioprinting for Infection and Transduction Studies" International Journal of Molecular Sciences 19, no. 10: 3129. https://doi.org/10.3390/ijms19103129
APA StyleHiller, T., Berg, J., Elomaa, L., Röhrs, V., Ullah, I., Schaar, K., Dietrich, A. -C., Al-Zeer, M. A., Kurtz, A., Hocke, A. C., Hippenstiel, S., Fechner, H., Weinhart, M., & Kurreck, J. (2018). Generation of a 3D Liver Model Comprising Human Extracellular Matrix in an Alginate/Gelatin-Based Bioink by Extrusion Bioprinting for Infection and Transduction Studies. International Journal of Molecular Sciences, 19(10), 3129. https://doi.org/10.3390/ijms19103129