Biological Roles of Ornithine Aminotransferase (OAT) in Plant Stress Tolerance: Present Progress and Future Perspectives
Abstract
:1. Introduction
2. Universality of OAT
2.1. General Kinetic Properties of the OAT Enzyme
2.2. Basic Similarities and Differences among Prokaryotes and Eukaryotes
3. OAT is Linked with Multiple Metabolic Pathways
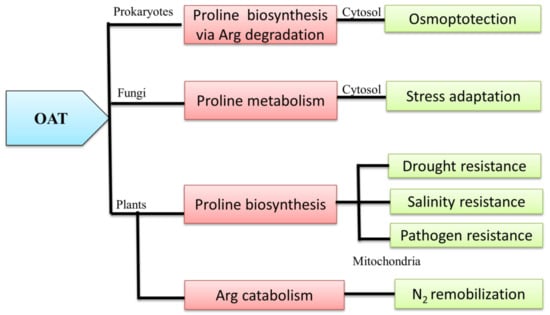
3.1. OAT and the Pro Metabolic Pathway
3.2. OAT is Involved in Arg Catabolism
3.2.1. Cyclic and Linear Orn Synthesis Pathways
3.2.2. Synthesis of Arg from Orn
4. Biological Roles of OAT
4.1. OAT is Involved in Stress-Induced Pro Accumulation
4.2. OAT Is Involved in Plant Non-Host Disease Resistance
4.3. Activation of Enzymes Involved in Stress-Induced Pro Accumulation
5. Future Directions
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ROS | reactive oxygen species |
| ABA | abscisic acid |
| OAT | ornithine amino transferase |
| P5CS | pyrroline-5-carboxylate synthase |
| P5C | pyrroline-5-carboxylate |
| P5CR | Pyrroline-5-carboxylate reductase |
| P5CDH | Pyrroline-5-carboxylate dehydrogenase |
| ProDH | Proline dehydrogenase |
| Orn | Ornithine |
| Glu | Glutamate |
| PCD | Programmed cell death |
| GSA | Glutamyl-5-semi-aldehyde |
| αKG | Alpha ketoglutarate |
| Pro | Proline |
| Arg | Arginine |
| ADI | Arg deiminase |
| Cit | Citrulline |
| HR | Hypersensitive |
| TFs | Transcription factors |
References
- Ramegowda, V.; Senthil-Kumar, M. The interactive effects of simultaneous biotic and abiotic stresses on plants: Mechanistic understanding from drought and pathogen combination. J. Plant Physiol. 2015, 176, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; De Vleesschauwer, D.; Sharma, M.K.; Ronald, P.C. Recent Advances in Dissecting Stress-Regulatory Crosstalk in Rice. Mol. Plant 2013, 6, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Nezhadahmadi, A.; Prodhan, Z.H.; Faruq, G. Drought Tolerance in Wheat. Sci. World J. 2013, 2013, 12. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Anwar, K.; Das, P.; Singla-Pareek, S.L.; Pareek, A. Overview of Methods for Assessing Salinity and Drought Tolerance of Transgenic Wheat Lines. In Wheat Biotechnology; Springer: New York, NY, USA, 2017; pp. 83–95. [Google Scholar]
- Das, G.; Patra, J.K.; Baek, K.-H. Insight into MAS: A Molecular Tool for Development of Stress Resistant and Quality of Rice through Gene Stacking. Front. Plant Sci. 2017, 8, 985. [Google Scholar] [CrossRef] [PubMed]
- McDonald, A.; Riha, S.; DiTommaso, A.; DeGaetano, A. Climate change and the geography of weed damage: Analysis of US maize systems suggests the potential for significant range transformations. Agric. Ecosyst. Environ. 2009, 130, 131–140. [Google Scholar] [CrossRef]
- Ziska, L.H.; Tomecek, M.B.; Gealy, D.R. Competitive interactions between cultivated and red rice as a function of recent and projected increases in atmospheric carbon dioxide. Agron. J. 2010, 102, 118–123. [Google Scholar] [CrossRef]
- Peters, K.; Breitsameter, L.; Gerowitt, B. Impact of climate change on weeds in agriculture: A review. Agron. Sustain. Dev. 2014, 34, 707–721. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.; Pandey, P.; Senthil-Kumar, M. Tailored responses to simultaneous drought stress and pathogen infection in plants. In Drought Stress Tolerance in Plants; Springer: New York, NY, USA, 2016; Volume 1, pp. 427–438. [Google Scholar]
- Ramu, V.S.; Paramanantham, A.; Ramegowda, V.; Mohan-Raju, B.; Udayakumar, M.; Senthil-Kumar, M. Transcriptome analysis of sunflower genotypes with contrasting oxidative stress tolerance reveals individual- and combined-biotic and abiotic stress tolerance mechanisms. PLoS ONE 2016, 11, e0157522. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Ramanarao, M.V.; Lee, S.; Kato, N.; Baisakh, N. Ectopic expression of ADP ribosylation factor 1 (SaARF1) from smooth cordgrass (Spartina alterniflora Loisel) confers drought and salt tolerance in transgenic rice and Arabidopsis. Plant Cell Tissue Organ Cult. 2014, 117, 17–30. [Google Scholar] [CrossRef]
- Vile, D.; Pervent, M.; Belluau, M.; Vasseur, F.; Bresson, J.; Muller, B.; Granier, C.; Simonneau, T. Arabidopsis growth under prolonged high temperature and water deficit: Independent or interactive effects? Plant Cell Environ. 2012, 35, 702–718. [Google Scholar] [CrossRef] [PubMed]
- Rizhsky, L.; Liang, H.; Mittler, R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002, 130, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Sainz, M.; Díaz, P.; Monza, J.; Borsani, O. Heat stress results in loss of chloroplast Cu/Zn superoxide dismutase and increased damage to Photosystem II in combined drought-heat stressed Lotus japonicus. Physiol. Plant. 2010, 140, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Rivero, R.M.; Mestre, T.C.; Mittler, R.; Rubio, F.; Garcia-sanchez, F.; Martinez, V. The combined effect of salinity and heat reveals a specific physiological, biochemical and molecular response in tomato plants. Plant Cell Environ. 2014, 37, 1059–1073. [Google Scholar] [CrossRef] [PubMed]
- Stránská, J.; Kopečný, D.; Tylichová, M.; Snégaroff, J.; Šebela, M. Ornithine δ-aminotransferase: An enzyme implicated in salt tolerance in higher plants. Plant Signal. Behav. 2008, 3, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Boon, L.; Geerts, W.J.; Jonker, A.; Lamers, W.H.; Van Noorden, C.J. High protein diet induces pericentral glutamate dehydrogenase and ornithine aminotransferase to provide sufficient glutamate for pericentral detoxification of ammonia in rat liver lobules. Histochem. Cell Biol. 1999, 111, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Ginguay, A.; Cynober, L.; Curis, E.; Nicolis, I. Ornithine Aminotransferase, an Important Glutamate-Metabolizing Enzyme at the Crossroads of Multiple Metabolic Pathways. Biology 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Krishna, R.V.; Leisinger, T. Biosynthesis of proline in Pseudomonas aeruginosa. Partial purification and characterization of γ-glutamyl kinase. Biochem. J. 1979, 181, 215–222. [Google Scholar] [PubMed]
- Csonka, L.N. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 1989, 53, 121–147. [Google Scholar] [PubMed]
- Adams, E.; Frank, L. Metabolism of proline and the hydroxyprolines. Annu. Rev. Biochem. 1980, 49, 1005–1061. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C. Proline accumulation: Biochemical aspects. In Physiology and Biochemistry of Drought Resistance in Plants; Academic Press: Sydney, Australia, 1981; pp. 243–359. [Google Scholar]
- Hu, C.; Delauney, A.J.; Verma, D. A bifunctional enzyme (Δ1-pyrroline-5-carboxylate synthetase) catalyzes the first two steps in proline biosynthesis in plants. Proc. Natl. Acad. Sci. USA 1992, 89, 9354–9358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.S.; Lu, Q.; Verma, D.P. Removal of feedback inhibition of Δ1-pyrroline-5-carboxylate synthetase, a bifunctional enzyme catalyzing the first two steps of proline biosynthesis in plants. J. Biol. Chem. 1995, 270, 20491–20496. [Google Scholar] [CrossRef] [PubMed]
- Perez-Arellano, I.; Carmona-Alvarez, F.; Gallego, J.; Cervera, J. Molecular mechanisms modulating glutamate kinase activity. Identification of the proline feedback inhibitor binding site. J. Mol. Biol. 2010, 404, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, F.; Cho, H.; Yelavarthi, V.; Sohn, C.; He, C.; Schneewind, O.; Bae, T. CcpA mediates proline auxotrophy and is required for Staphylococcus aureus pathogenesis. J. Bacteriol. 2010, 192, 3883–3892. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-D. Pathways and regulation of bacterial arginine metabolism and perspectives for obtaining arginine overproducing strains. Appl. Microbiol. Biotechnol. 2006, 70, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Teng, J.L.L.; Botelho, M.G.; Lo, R.C.; Lau, S.K.P.; Woo, P.C.Y. Arginine Metabolism in Bacterial Pathogenesis and Cancer Therapy. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [Green Version]
- Hampel, A.; Huber, C.; Geffers, R.; Spona-Friedl, M.; Eisenreich, W.; Bange, F.-C. Mycobacterium tuberculosis is a natural ornithine aminotransferase (rocD) mutant and depends on Rv2323c for growth on arginine. PLoS ONE 2015, 10, e0136914. [Google Scholar]
- Fuhrmann, J.; Thompson, P.R. Protein Arginine Methylation and Citrullination in. ACS Chem. Biol. 2016, 11, 654–668. [Google Scholar] [PubMed]
- Fincham, J. Ornithine transaminase in Neurospora and its relation to the biosynthesis of proline. Biochem. J. 1953, 53, 313. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.L. Intracellular localization of ornithine and arginine pools in Neurospora. J. Biol. Chem. 1973, 248, 5409–5413. [Google Scholar] [PubMed]
- Jauniaux, J.-C.; Urrestarazu, L.A.; Wiame, J.-M. Arginine metabolism in Saccharomyces cerevisiae: Subcellular localization of the enzymes. J. Bacteriol. 1978, 133, 1096–1107. [Google Scholar] [PubMed]
- Wagemaker, M.J.; Eastwood, D.C.; Welagen, J.; van der Drift, C.; Jetten, M.S.; Burton, K.; Van Griensven, L.J.; Op den Camp, H.J. The role of ornithine aminotransferase in fruiting body formation of the mushroom Agaricus bisporus. Mycol. Res. 2007, 111, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Gafan, C.; Wilson, J.; Berger, L.C.; Berger, B.J. Characterization of the ornithine aminotransferase from Plasmodium falciparum. Mol. Biochem. Parasitol. 2001, 118, 1–10. [Google Scholar] [CrossRef]
- Dzikowska, A.; Swianiewicz, M.; Talarczyk, A.; Wisniewska, M.; Goras, M.; Scazzocchio, C.; Weglenski, P. Cloning, characterisation and regulation of the ornithine transaminase (otaA) gene of Aspergillus nidulans. Curr. Genet. 1999, 35, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Scher, W.I.; Vogel, H.J. Occurrence of ornithine delta-transaminase: A dichotomy. Proc. Natl. Acad. Sci. USA 1957, 43, 796–803. [Google Scholar] [CrossRef] [PubMed]
- Bone, D.H. Metabolism of Citrulline and Ornithine in Mung Bean Mitochondria. Plant Physiol. 1959, 34, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Mazelis, M.; Fowden, L. Conversion of ornithine into proline by enzymes from germinating peanut cotyledons. Phytochemistry 1969, 8, 801–809. [Google Scholar] [CrossRef]
- Splittstoesser, W.; Fowden, L. Ornithine transaminase from Cucurbita maxima cotyledons. Phytochemistry 1973, 12, 785–790. [Google Scholar] [CrossRef]
- Lu, T.S.; Mazelis, M. l-Ornithine:2-Oxoacid Aminotransferase from Squash (Cucurbita pepo, L.) Cotyledons: Purification and Properties. Plant Physiol. 1975, 55, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Delauney, A.; Hu, C.; Kishor, P.; Verma, D. Cloning of ornithine delta-aminotransferase cDNA from Vigna aconitifolia by trans-complementation in Escherichia coli and regulation of proline biosynthesis. J. Biol. Chem. 1993, 268, 18673–18678. [Google Scholar] [PubMed]
- Roosens, N.H.; Thu, T.T.; Iskandar, H.M.; Jacobs, M. Isolation of the ornithine-δ-aminotransferase cDNA and effect of salt stress on its expression in Arabidopsis thaliana. Plant Physiol. 1998, 117, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Funck, D.; Stadelhofer, B.; Koch, W. Ornithine-delta-aminotransferase is essential for arginine catabolism but not for proline biosynthesis. BMC Plant Biol 2008, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Strizhov, N.; Abraham, E.; Okresz, L.; Blickling, S.; Zilberstein, A.; Schell, J.; Koncz, C.; Szabados, L. Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 1997, 12, 557–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savouré, A.; Jaoua, S.; Hua, X.-J.; Ardiles, W.; Van Montagu, M.; Verbruggen, N. Isolation, characterization, and chromosomal location of a gene encoding the Δ1-pyrroline-5-carboxylate synthetase in Arabidopsis thaliana. FEBS Lett. 1995, 372, 13–19. [Google Scholar] [CrossRef]
- Szoke, A.; Miao, G.-H.; Hong, Z.; Verma, D.P.S. Subcellular location of δ1-pyrroline-5-carboxylate reductase in root/nodule and leaf of soybean. Plant Physiol. 1992, 99, 1642–1649. [Google Scholar] [CrossRef] [PubMed]
- Verbruggen, N.; Villarroel, R.; Van Montagu, M. Osmoregulation of a pyrroline-5-carboxylate reductase gene in Arabidopsis thaliana. Plant Physiol. 1993, 103, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, C.; Pizzuto, R.; Pallotta, M.L.; De Santis, A.; Passarella, S. Mitochondrial transport in proline catabolism in plants: The existence of two separate translocators in mitochondria isolated from durum wheat seedlings. Planta 2006, 223, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Szekely, G.; Abraham, E.; Cseplo, A.; Rigo, G.; Zsigmond, L.; Csiszar, J.; Ayaydin, F.; Strizhov, N.; Jasik, J.; Schmelzer, E.; et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008, 53, 11–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verslues, P.E.; Sharma, S. Proline Metabolism and Its Implications for Plant-Environment Interaction. Arabidopsis Book/Am. Soc. Plant Biol. 2010, 8, e0140. [Google Scholar] [CrossRef] [PubMed]
- Senthil-Kumar, M.; Mysore, K.S. Ornithine-delta-aminotransferase and proline dehydrogenase genes play a role in non-host disease resistance by regulating pyrroline-5-carboxylate metabolism-induced hypersensitive response. Plant Cell Environ. 2012, 35, 1329–1343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Planchais, S.; Cabassa, C.; Toka, I.; Justin, A.-M.; Renou, J.-P.; Savouré, A.; Carol, P. BASIC AMINO ACID CARRIER 2 gene expression modulates arginine and urea content and stress recovery in Arabidopsis leaves. Front. Plant Sci. 2014, 5, 330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slocum, R.D. Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol. Biochem. 2005, 43, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Szabados, L.; Savoure, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef] [PubMed]
- Witte, C.-P. Urea metabolism in plants. Plant Sci. 2011, 180, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Honig, A.; Stein, H.; Suzuki, N.; Mittler, R.; Zilberstein, A. Unraveling Δ1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J. Biol. Chem. 2009, 284, 26482–26492. [Google Scholar] [CrossRef] [PubMed]
- Hervieu, F.; Dily, F.; Huault, C.; BILLARD, J.P. Contribution of ornithine aminotransferase to proline accumulation in NaCl-treated radish cotyledons. Plant Cell Environ. 1995, 18, 205–210. [Google Scholar] [CrossRef]
- Yang, C.-W.; Kao, C.H. Importance of ornithine-δ-aminotransferase to proline accumulation caused by water stress in detached rice leaves. Plant Growth Regul. 1999, 27, 191–194. [Google Scholar] [CrossRef]
- Roosens, N.H.; Bitar, F.A.; Loenders, K.; Angenon, G.; Jacobs, M. Overexpression of ornithine-δ-aminotransferase increases proline biosynthesis and confers osmotolerance in transgenic plants. Mol. Breed. 2002, 9, 73–80. [Google Scholar] [CrossRef]
- Wu, L.; Fan, Z.; Guo, L.; Li, Y.; Zhang, W.; Qu, L.-J.; Chen, Z. Over-expression of an Arabidopsis δ-OAT gene enhances salt and drought tolerance in transgenic rice. Chin. Sci. Bull. 2003, 48, 2594–2600. [Google Scholar] [CrossRef]
- Armengaud, P.; Thiery, L.; Buhot, N.; Grenier-de March, G.; Savouré, A. Transcriptional regulation of proline biosynthesis in Medicago truncatula reveals developmental and environmental specific features. Physiol. Plant. 2004, 120, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Verslues, P.E. Mechanisms independent of abscisic acid (ABA) or proline feedback have a predominant role in transcriptional regulation of proline metabolism during low water potential and stress recovery. Plant Cell Environ. 2010, 33, 1838–1851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, J.; Hu, H.; Xiong, L. An ornithine δ-aminotransferase gene OsOAT confers drought and oxidative stress tolerance in rice. Plant Sci. 2012, 197, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; You, J.; Fang, Y.; Zhu, X.; Qi, Z.; Xiong, L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2008, 67, 169–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, X.; Zhang, L.; Natarajan, S.K.; Becker, D.F. Proline mechanisms of stress survival. Antioxid. Redox Signal. 2013, 19, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Dougall, D.K.; Fulton, M.M. Biosynthesis of protein amino acids in plant tissue culture. III. Studies on the biosynthesis of arginine. Plant Physiol. 1967, 42, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Cunin, R.; Glansdorff, N.; Pierard, A.; Stalon, V. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 1986, 50, 314. [Google Scholar] [PubMed]
- Davis, R.H. Compartmental and regulatory mechanisms in the arginine pathways of Neurospora crassa and Saccharomyces cerevisiae. Microbiol. Rev. 1986, 50, 280. [Google Scholar] [PubMed]
- Shargool, D.; Jain, J.; McKay, G. Ornithine biosynthesis, and arginine biosynthesis and degradation in plant cells. Phytochemistry 1988, 27, 1571–1574. [Google Scholar] [CrossRef]
- Vogel, H.J.; Bonner, D.M. Acetylornithinase of Escherichia coli: Partial purification and some properties. J. Biol. Chem. 1956, 218, 97–106. [Google Scholar] [PubMed]
- Meinnel, T.; Schmitt, E.; Mechulam, Y.; Blanquet, S. Structural and biochemical characterization of the Escherichia coli argE gene product. J. Bacteriol. 1992, 174, 2323–2331. [Google Scholar] [CrossRef] [PubMed]
- Crabeel, M.; Abadjieva, A.; Hilven, P.; Desimpelaere, J.; Soetens, O. Characterization of the Saccharomyces cerevisiae ARG7 gene encoding ornithine acetyltransferase, an enzyme also endowed with acetylglutamate synthase activity. Eur. J. Biochem. 1997, 250, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Page, A.F.; Minocha, R.; Minocha, S.C. Living with high putrescine: Expression of ornithine and arginine biosynthetic pathway genes in high and low putrescine producing poplar cells. Amino Acids 2012, 42, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Frémont, N.; Riefler, M.; Stolz, A.; Schmülling, T. The Arabidopsis TUMOR PRONE5 gene encodes an acetylornithine aminotransferase required for arginine biosynthesis and root meristem maintenance in blue light. Plant Physiol. 2013, 161, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Mennella, G.; Martini, F.; Francese, G.; Pandolfini, T. Involvement of the putative N-acetylornithine deacetylase from Arabidopsis thaliana in flowering and fruit development. Plant Cell Physiol. 2015, pcv030. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.J.; Thompson, J.F.; Johnson, C.M. Metabolism of glutamic acid and N-acetylglutamic acid in leaf discs and cell-free extracts of higher plants. Plant Physiol. 1969, 44, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Caldovic, L.; Tuchman, M. N-acetylglutamate and its changing role through evolution. Biochem. J. 2003, 372, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Micallef, B.J.; Shelp, B.J. Arginine metabolism in developing soybean cotyledons I. Relationship to nitrogen nutrition. Plant Physiol. 1989, 90, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, S.; Miyake, C.; Kohchi, T.; Fujii, S.; Uchida, M.; Yokota, A. Responses of wild watermelon to drought stress: Accumulation of an ArgE homologue and citrulline in leaves during water deficits. Plant Cell Physiol. 2000, 41, 864–873. [Google Scholar] [CrossRef] [PubMed]
- Kusvuran, S.; Dasgan, H.Y.; Abak, K. Citrulline Is an Important Biochemical Indicator in Tolerance to Saline and Drought Stresses in Melon. Sci. World J. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Goldraij, A.; Polacco, J.C. Arginine degradation by arginase in mitochondria of soybean seedling cotyledons. Planta 2000, 210, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Qamar, A.; Mysore, K.; Senthil-Kumar, M. Role of proline and pyrroline-5-carboxylate metabolism in plant defense against invading pathogens. Front. Plant Sci. 2015, 6, 503. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Liu, A.; Hua, X. Proline accumulation and transcriptional regulation of proline biosynthesis and degradation in Brassica napus. BMB Rep. 2009, 42, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Kemble, A.; Macpherson, H.T. Liberation of amino acids in perennial rye grass during wilting. Biochem. J. 1954, 58, 46. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, N.; Sairam, R.; Tyagi, A. Expression of Δ1-pyrroline-5-carboxylate synthetase gene during drought in rice (Oryza sativa L.). Indian J. Biochem. Biophys. 2005, 42, 366–370. [Google Scholar] [PubMed]
- Yang, S.-L.; Lan, S.-S.; Gong, M. Hydrogen peroxide-induced proline and metabolic pathway of its accumulation in maize seedlings. J. Plant Physiol. 2009, 166, 1694–1699. [Google Scholar] [CrossRef] [PubMed]
- Yoshiba, Y.; Kiyosue, T.; Katagiri, T.; Ueda, H.; Mizoguchi, T.; Yamaguchi-Shinozaki, K.; Wada, K.; Harada, Y.; Shinozaki, K. Correlation between the induction of a gene for Δ1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J. 1995, 7, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Saradhi, P.P.; AliaArora, S.; Prasad, K. Proline accumulates in plants exposed to UV radiation and protects them against UV-induced peroxidation. Biochem. Biophys. Res. Commun. 1995, 209, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Schat, H.; Sharma, S.S.; Vooijs, R. Heavy metal-induced accumulation of free proline in a metal-tolerant and a nontolerant ecotype of Silene vulgaris. Physiol. Plant. 1997, 101, 477–482. [Google Scholar] [CrossRef]
- Fabro, G.; Kovács, I.; Pavet, V.; Szabados, L.; Alvarez, M.E. Proline accumulation and AtP5CS2 gene activation are induced by plant-pathogen incompatible interactions in Arabidopsis. Mol. Plant-Microbe Interact. 2004, 17, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Haudecoeur, E.; Planamente, S.; Cirou, A.; Tannieres, M.; Shelp, B.; Morera, S.; Faure, D. Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 2009, 106, 14587–14592. [Google Scholar] [CrossRef] [PubMed]
- Csonka, L.; Gelvin, S.; Goodner, B.; Orser, C.; Siemieniak, D.; Slightom, J. Nucleotide sequence of a mutation in the proB gene of Escherichia coli that confers proline overproduction and enhanced tolerance to osmotic stress. Gene 1988, 64, 199–205. [Google Scholar] [CrossRef]
- Csonka, L.N.; Hanson, A.D. Prokaryotic osmoregulation: Genetics and physiology. Annu. Rev. Microbiol. 1991, 45, 569–606. [Google Scholar] [CrossRef] [PubMed]
- Da Rocha, I.M.A.; Vitorello, V.A.; Silva, J.S.; Ferreira-Silva, S.L.; Viégas, R.A.; Silva, E.N.; Silveira, J.A.G. Exogenous ornithine is an effective precursor and the δ-ornithine amino transferase pathway contributes to proline accumulation under high N recycling in salt-stressed cashew leaves. J. Plant Physiol. 2012, 169, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Canas, R.A.; Villalobos, D.P.; Diaz-Moreno, S.M.; Canovas, F.M.; Canton, F.R. Molecular and functional analyses support a role of Ornithine-δ-aminotransferase in the provision of glutamate for glutamine biosynthesis during pine germination. Plant Physiol. 2008, 148, 77–88. [Google Scholar] [CrossRef] [PubMed]
- She, M.; Wang, J.; Wang, X.; Yin, G.; Wang, K.; Du, L.; Ye, X. Comprehensive molecular analysis of arginase-encoding genes in common wheat and its progenitor species. Sci. Rep. 2017, 7, 6641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Xue, Z.; Tang, D.; Shen, Y.; Shi, W.; Ren, L.; Du, G.; Li, Y.; Cheng, Z. Ornithine δ-aminotransferase is critical for floret development and seed setting through mediating nitrogen reutilization in rice. Plant J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, H.; Funck, D.; Rentsch, D.; Frommer, W.B. Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous proline application. Plant Physiol. 2000, 122, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Deuschle, K.; Funck, D.; Forlani, G.; Stransky, H.; Biehl, A.; Leister, D.; van der Graaff, E.; Kunze, R.; Frommer, W.B. The role of Δ1-pyrroline-5-carboxylate dehydrogenase in proline degradation. Plant Cell 2004, 16, 3413–3425. [Google Scholar] [CrossRef] [PubMed]
- Amirsadeghi, S.; Robson, C.A.; Vanlerberghe, G.C. The role of the mitochondrion in plant responses to biotic stress. Physiol. Plant. 2007, 129, 253–266. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Mysore, K.S. Nonhost resistance against bacterial pathogens: Retrospectives and prospects. Annu. Rev. Phytopathol. 2013, 51, 407–427. [Google Scholar] [CrossRef] [PubMed]
- Kishor, P.K.; Sangam, S.; Amrutha, R.; Laxmi, P.S.; Naidu, K.; Rao, K.; Rao, S.; Reddy, K.; Theriappan, P.; Sreenivasulu, N. Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: Its implications in plant growth and abiotic stress tolerance. Curr. Sci. 2005, 88, 424–438. [Google Scholar]
- Verbruggen, N.; Hermans, C. Proline accumulation in plants: A review. Amino Acids 2008, 35, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Monteoliva, M.I.; Rizzi, Y.S.; Cecchini, N.M.; Hajirezaei, M.-R.; Alvarez, M.E. Context of action of proline dehydrogenase (ProDH) in the hypersensitive response of Arabidopsis. BMC Plant Biol. 2014, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Cecchini, N.M.; Monteoliva, M.I.; Alvarez, M.E. Proline dehydrogenase contributes to pathogen defense in Arabidopsis. Plant Physiol. 2011, 155, 1947–1959. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, A.; Nasuno, R.; Takagi, H. The proline metabolism intermediate Δ1-pyrroline-5-carboxylate directly inhibits the mitochondrial respiration in budding yeast. FEBS Lett. 2012, 586, 2411–2416. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.R.; Lui, E.Y.; Chow, E.W.; Arras, S.D.; Morrow, C.A.; Fraser, J.A. Reactive oxygen species homeostasis and virulence of the fungal pathogen Cryptococcus neoformans requires an intact proline catabolism pathway. Genetics 2013, 194, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Rizzi, Y.; Monteoliva, M.; Fabro, G.; Grosso, C.; Laróvere, L.; Alvarez, M. P5CDH affects the pathways contributing to Pro synthesis after ProDH activation by biotic and abiotic stress conditions. Front. Plant Sci. 2015, 6, 572. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.M.; Thomas, V.; Bennett, M.H.; Mansfield, J.; Grant, M. Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 2006, 142, 1603–1620. [Google Scholar] [CrossRef] [PubMed]
- Fichman, Y.; Gerdes, S.Y.; Kovács, H.; Szabados, L.; Zilberstein, A.; Csonka, L.N. Evolution of proline biosynthesis: Enzymology, bioinformatics, genetics, and transcriptional regulation. Biol. Rev. 2015, 90, 1065–1099. [Google Scholar] [CrossRef] [PubMed]
- KISHOR, K.; Polavarapu, B.; Sreenivasulu, N. Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ. 2014, 37, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Weltmeier, F.; Ehlert, A.; Mayer, C.S.; Dietrich, K.; Wang, X.; Schutze, K.; Alonso, R.; Harter, K.; Vicente-Carbajosa, J.; Droge-Laser, W. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J. 2006, 25, 3133–3143. [Google Scholar] [CrossRef] [PubMed]
- Kiyosue, T.; Yoshiba, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell 1996, 8, 1323–1335. [Google Scholar] [CrossRef] [PubMed]
- Hare, P.; Cress, W. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul. 1997, 21, 79–102. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Golldack, D.; Luking, I.; Yang, O. Plant tolerance to drought and salinity: Stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011, 30, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Udvardi, M.K.; Kakar, K.; Wandrey, M.; Montanari, O.; Murray, J.; Andriankaja, A.; Zhang, J.Y.; Benedito, V.; Hofer, J.M.; Chueng, F.; et al. Legume transcription factors: Global regulators of plant development and response to the environment. Plant Physiol. 2007, 144, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, L.; Di, Z.; Yang, W.; Liu, J.; Li, M.; Wang, X.; Cui, C.; Wang, X.; Wang, X.; Zhang, R.; et al. Overexpression of ERF1-V from Haynaldia villosa Can Enhance the Resistance of Wheat to Powdery Mildew and Increase the Tolerance to Salt and Drought Stresses. Front Plant Sci 2017, 8, 1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarattini, M.; Forlani, G. Toward Unveiling the Mechanisms for Transcriptional Regulation of Proline Biosynthesis in the Plant Cell Response to Biotic and Abiotic Stress Conditions. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco-Zorrilla, J.M.; Solano, R. Identification of plant transcription factor target sequences. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Ohad, N.; Yalovsky, S. Utilizing bimolecular fluorescence complementation (BiFC) to assay protein-protein interaction in plants. Methods Mol. Biol. 2010, 655, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Gao, C. Targeted genome modification technologies and their applications in crop improvements. Plant Cell Rep. 2014, 33, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Riaz, B.; Ye, X. Wheat genome editing expedited by efficient transformation techniques: Progress and perspectives. Crop J. 2017. [Google Scholar] [CrossRef]





| No. | Name | Accession Number | No. | Name | Accession Number |
|---|---|---|---|---|---|
| 1 | Arabidopsis thaliana | OAO92185 | 20 | Capsella rubella | XP_006280404 |
| 2 | Vitis vinifera | NP_001268069 | 21 | Camelina sativa | XP_010494787 |
| 3 | Medicago truncatula | Q8GUA8 | 22 | Ricinus communis | EEF42620 |
| 4 | Oryza sativa | XP_015630389.1 | 23 | Jatropha curcas | NP_001306851 |
| 5 | Glycine max | XP_003531161.1 | 24 | Populus euphratica | XP_011007419 |
| 6 | Sorghum bicolor | XP_002464174.1 | 25 | Gossypium hirsutum | XP_016753478 |
| 7 | Zea mays | NP_001130350.1 | 26 | Gossypium arboreum | XP_017641965 |
| 8 | Ricinus communis | XP_002519647.2 | 27 | Gossypium raimondii | XP_012450413 |
| 9 | Helianthus tuberosus | AHJ08571.1 | 28 | Prunus persica | XP_007214014 |
| 10 | Nicotiana attenuata | XP_019259981.1 | 29 | Rosa chinensis | XP_024156782 |
| 11 | Brassica napus | NP_001303219.1 | 30 | Carica papaya | XP_021904606 |
| 12 | Brassica oleracea | XP_013593040.1 | 31 | Cucurbita maxima | XP_023001421 |
| 13 | Brassica rapa | NP_001288848.1 | 32 | Solanum tuberosum | XP_006355410 |
| 14 | Hordeum vulgare | BAJ87243.1 | 33 | Solanum lycopersicum | XP_015085834 |
| 15 | Aquilegia coerulea | PIA41644.1 | 34 | Eutrema salsugineum | XP_006398303 |
| 16 | Nicotiana attenuata | XP_019259981.1 | 35 | Cucurbita maxima | XP_022994797.1 |
| 17 | Nicotiana tabacum | XP_016456334.1 | 36 | Capsicum chinense | PHU09018.1 |
| 18 | Prunus persica | ALT55650.1 | 37 | Capsicum annuum | XP_016537501.1 |
| 19 | Ziziphus jujuba | XP_009775369.1 | 38 | Sesamum indicum | XP_011096597.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anwar, A.; She, M.; Wang, K.; Riaz, B.; Ye, X. Biological Roles of Ornithine Aminotransferase (OAT) in Plant Stress Tolerance: Present Progress and Future Perspectives. Int. J. Mol. Sci. 2018, 19, 3681. https://doi.org/10.3390/ijms19113681
Anwar A, She M, Wang K, Riaz B, Ye X. Biological Roles of Ornithine Aminotransferase (OAT) in Plant Stress Tolerance: Present Progress and Future Perspectives. International Journal of Molecular Sciences. 2018; 19(11):3681. https://doi.org/10.3390/ijms19113681
Chicago/Turabian StyleAnwar, Alia, Maoyun She, Ke Wang, Bisma Riaz, and Xingguo Ye. 2018. "Biological Roles of Ornithine Aminotransferase (OAT) in Plant Stress Tolerance: Present Progress and Future Perspectives" International Journal of Molecular Sciences 19, no. 11: 3681. https://doi.org/10.3390/ijms19113681
APA StyleAnwar, A., She, M., Wang, K., Riaz, B., & Ye, X. (2018). Biological Roles of Ornithine Aminotransferase (OAT) in Plant Stress Tolerance: Present Progress and Future Perspectives. International Journal of Molecular Sciences, 19(11), 3681. https://doi.org/10.3390/ijms19113681