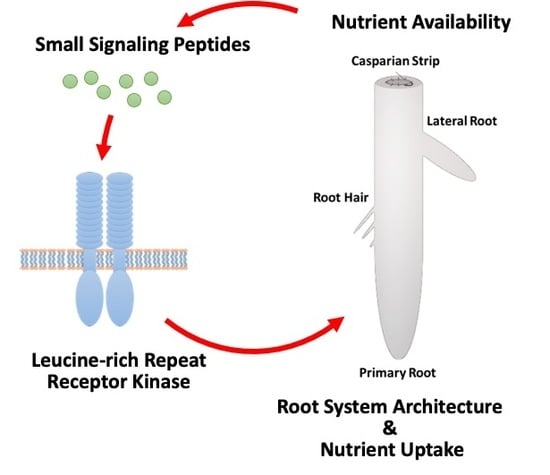
Nutrient-Responsive Small Signaling Peptides and Their Influence on the Root System Architecture
Abstract
:1. Introduction
2. Nitrogen-Responsive SSPs
3. Phosphorus-Responsive SSPs
4. SSP-Dependent Changes on RSA Have Impact on Nutrient Homeostasis
5. Transcriptomic Studies Reveal Potential SSP Candidates for Functional Characterization
6. Gene Expression Reveals Potential for Nutrient-Dependent Crosstalk Involving SSP Pathways
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| RSA | Root system architecture |
| SSP | Small signaling peptide |
| LRR-RK | Leucine-rich repeat receptor kinase |
| sORF | Small open reading frames |
| NRT | Nitrate transporter |
| CLE | Clavata3/embryo surrounding region related |
| AON | Autoregulation of nodulation |
| CLV | CLAVATA |
| CEP | C-terminally encoded peptide |
| RGF/GLV/CLEL | Root growth factor/Golven/CLE-like |
| LPR | Low phosphate root |
| PDR | Phosphate deficiency response |
| PLT | PLETHORA |
| IDA | Inflorescence deficient in abscission |
| PSY | Plant peptide containing sulfated tyrosine |
| PIP | PAMP-induced secreted peptide |
| CIF | Casparian strip integrity factor |
| GSO | GASSHO1/SCHENGEN3 |
| RALF | Rapid alkalization factor |
| FER | FERONIA |
| HAE | HAESA |
| PSK | PHYTOSULFOKINE |
| CAPE | Cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 protein |
| NMR | Nuclear magnetic resonance |
| LC-MS/MS | Liquid chromatography with tandem mass spectrometry |
| BAM | Barely any meristem |
| CRN | CORYNE |
| RPK | Receptor-like protein kinase |
| SERK | Somatic embryogenesis receptor-like kinases |
References
- Giehl, R.F.H.; von Wiren, N. Root nutrient foraging. Plant Physiol. 2014, 166, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Osmont, K.S.; Sibout, R.; Hardtke, C.S. Hidden branches: Developments in root system architecture. Annu. Rev. Plant Biol. 2007, 58, 93–113. [Google Scholar] [CrossRef] [PubMed]
- Gruber, B.D.; Giehl, R.F.H.; Friedel, S.; von Wirén, N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013, 163, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, Z.; Amtmann, A. Food for thought: How nutrients regulate root system architecture. Curr. Opin. Plant Biol. 2017, 39, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Tavormina, P.; Coninck, B.D.; Nikonorova, N.; Smet, I.D.; Cammue, B.P.A. The plant peptidome: An expanding repertoire of structural features and biological functions. Plant Cell 2015, 27, 2095–2118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czyzewicz, N.; Smet, I.D. The Arabidopsis thaliana CLAVATA3/EMBRYO-SURROUNDING REGION 26 (CLE26) peptide is able to alter root architecture of Solanum lycopersicum and Brassica napus. Plant Signal. Behav. 2016, 11, e1118598. [Google Scholar] [CrossRef] [PubMed]
- Van de Velde, W.; Zehirov, G.; Szatmari, A.; Debreczeny, M.; Ishihara, H.; Kevei, Z.; Farkas, A.; Mikulass, K.; Nagy, A.; Tiricz, H.; et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 2010, 327, 1122–1126. [Google Scholar] [CrossRef]
- Shiu, S.-H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [Green Version]
- Matsubayashi, Y. Post-translational modifications in secreted peptide hormones in plants. Plant Cell Physiol. 2011, 52, 5–13. [Google Scholar] [CrossRef]
- Hobe, M.; Müller, R.; Grünewald, M.; Brand, U.; Simon, R. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev. Genes Evol. 2003, 213, 371–381. [Google Scholar] [CrossRef]
- Suzaki, T.; Yoshida, A.; Hirano, H.-Y. Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice. Plant Cell 2008, 20, 2049–2058. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, M.; Yamaguchi, T.; Kazama, T.; Ito, T.; Horiguchi, G.; Tsukaya, H. ROTUNDIFOLIA4 regulates cell proliferation along the body axis in arabidopsis shoot. Plant Cell Physiol. 2011, 52, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-L.; Xie, L.-F.; Mao, H.-Z.; Puah, C.S.; Yang, W.-C.; Jiang, L.; Sundaresan, V.; Ye, D. TAPETUM DETERMINANT1 is required for cell specialization in the Arabidopsis anther. Plant Cell 2003, 15, 2792–2804. [Google Scholar] [CrossRef] [PubMed]
- Araya, T.; Miyamoto, M.; Wibowo, J.; Suzuki, A.; Kojima, S.; Tsuchiya, Y.N.; Sawa, S.; Fukuda, H.; von Wirén, N.; Takahashi, H. CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 2029–2034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumpf, R.P.; Shi, C.-L.; Larrieu, A.; Stø, I.M.; Butenko, M.A.; Péret, B.; Riiser, E.S.; Bennett, M.J.; Aalen, R.B. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. USA 2013, 201210835. [Google Scholar] [CrossRef] [PubMed]
- Mortier, V.; Herder, G.D.; Whitford, R.; de Velde, W.V.; Rombauts, S.; D’haeseleer, K.; Holsters, M.; Goormachtig, S. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 2010, 153, 222–237. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.; Drozdzecki, A.; Hoogewijs, K.; Nguyen, A.; Beeckman, T.; Madder, A.; Hilson, P. Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation. Plant Physiol. 2013, 161, 954–970. [Google Scholar] [CrossRef]
- Lynch, J.P. Steep, cheap and deep: An ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef]
- Remans, T.; Nacry, P.; Pervent, M.; Filleur, S.; Diatloff, E.; Mounier, E.; Tillard, P.; Forde, B.G.; Gojon, A. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA 2006, 103, 19206–19211. [Google Scholar] [CrossRef] [Green Version]
- Lima, J.E.; Kojima, S.; Takahashi, H.; Wirén, N. von Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. Plant Cell 2010, 22, 3621–3633. [Google Scholar] [CrossRef]
- Canales, J.; Contreras-López, O.; Álvarez, J.M.; Gutiérrez, R.A. Nitrate induction of root hair density is mediated by TGA1/TGA4 and CPC transcription factors in Arabidopsis thaliana. Plant J. 2017, 92, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Cock, J.M.; McCormick, S. A large family of genes that share homology with CLAVATA3. Plant Physiol. 2001, 126, 939–942. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Shinohara, H.; Mori, T.; Matsubayashi, Y.; Kawaguchi, M. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 2013, 4, 2191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brand, U.; Fletcher, J.C.; Hobe, M.; Meyerowitz, E.M.; Simon, R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 2000, 289, 617–619. [Google Scholar] [CrossRef] [PubMed]
- Stahl, Y.; Wink, R.H.; Ingram, G.C.; Simon, R. A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 2009, 19, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Scheible, W.-R.; Morcuende, R.; Czechowski, T.; Fritz, C.; Osuna, D.; Palacios-Rojas, N.; Schindelasch, D.; Thimm, O.; Udvardi, M.K.; Stitt, M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004, 136, 2483–2499. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, K.; Ogawa, M.; Matsubayashi, Y. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 2008, 55, 152–160. [Google Scholar] [CrossRef] [Green Version]
- Delay, C.; Imin, N.; Djordjevic, M.A. CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. J. Exp. Bot. 2013, 64, 5383–5394. [Google Scholar] [CrossRef] [Green Version]
- Ohkubo, Y.; Tanaka, M.; Tabata, R.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 2017, 3, 17029. [Google Scholar] [CrossRef]
- Tabata, R.; Sumida, K.; Yoshii, T.; Ohyama, K.; Shinohara, H.; Matsubayashi, Y. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 2014, 346, 343–346. [Google Scholar] [CrossRef]
- Roberts, I.; Smith, S.; Stes, E.; De Rybel, B.; Staes, A.; van de Cotte, B.; Njo, M.F.; Dedeyne, L.; Demol, H.; Lavenus, J.; et al. CEP5 and XIP1/CEPR1 regulate lateral root initiation in Arabidopsis. J. Exp. Bot. 2016, 67, 4889–4899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imin, N.; Mohd-Radzman, N.A.; Ogilvie, H.A.; Djordjevic, M.A. The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J. Exp. Bot. 2013, 64, 5395–5409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohd-Radzman, N.A.; Laffont, C.; Ivanovici, A.; Patel, N.; Reid, D.E.; Stougaard, J.; Frugier, F.; Imin, N.; Djordjevic, M.A. Different pathways act downstream of the peptide receptor CRA2 to regulate lateral root and nodule development. Plant Physiol. 2016, 171, 2536–2548. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, M.A.; Mohd-Radzman, N.A.; Imin, N. Small-peptide signals that control root nodule number, development, and symbiosis. J. Exp. Bot. 2015, 66, 5171–5181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, J.P.; Brown, K.M. Topsoil foraging—An architectural adaptation of plants to low phosphorus availability. Plant Soil 2001, 237, 225–237. [Google Scholar] [CrossRef]
- Linkohr, B.I.; Williamson, L.C.; Fitter, A.H.; Leyser, H.M.O. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 2002, 29, 751–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ticconi, C.A.; Lucero, R.D.; Sakhonwasee, S.; Adamson, A.W.; Creff, A.; Nussaume, L.; Desnos, T.; Abel, S. ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc. Natl. Acad. Sci. USA 2009, 106, 14174–14179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deb, S.; Sankaranarayanan, S.; Wewala, G.; Widdup, E.; Samuel, M.A. The S-domain receptor kinase Arabidopsis Receptor Kinase2 and the U box/armadillo repeat-containing E3 ubiquitin ligase9 module mediates lateral root development under phosphate starvation in Arabidopsis. Plant Physiol. 2014, 165, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Yu, H.; Dong, J.; Che, X.; Jiao, Y.; Liu, D. The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet. 2016, 12, e1006194. [Google Scholar] [CrossRef]
- Cederholm, H.M.; Benfey, P.N. Distinct sensitivities to phosphate deprivation suggest that RGF peptides play disparate roles in Arabidopsis thaliana root development. New Phytol. 2015, 207, 683–691. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, Y.; Ogawa-Ohnishi, M.; Mori, A.; Matsubayashi, Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 2010, 329, 1065–1067. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, H.; Mori, A.; Yasue, N.; Sumida, K.; Matsubayashi, Y. Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 3897–3902. [Google Scholar] [CrossRef] [PubMed]
- de Bang, T.C.; Lundquist, P.K.; Dai, X.; Boschiero, C.; Zhuang, Z.; Pant, P.; Torres-Jerez, I.; Roy, S.; Nogales, J.; Veerappan, V.; et al. Genome-Wide Identification of Medicago peptides involved in macronutrient responses and nodulation. Plant Physiol. 2017, 175, 1669–1689. [Google Scholar] [CrossRef] [PubMed]
- Doblas, V.G.; Smakowska-Luzan, E.; Fujita, S.; Alassimone, J.; Barberon, M.; Madalinski, M.; Belkhadir, Y.; Geldner, N. Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 2017, 355, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Shinohara, H.; Tanaka, M.; Baba, K.; Ogawa-Ohnishi, M.; Matsubayashi, Y. A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 2017, 355, 284–286. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.E.; Trontin, C.; Duan, L.; Dinneny, J.R. Beyond the barrier: Communication in the root through the endodermis. Plant Physiol. 2014, 166, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef]
- Wu, J.; Kurten, E.L.; Monshausen, G.; Hummel, G.M.; Gilroy, S.; Baldwin, I.T. NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. Plant J. 2007, 52, 877–890. [Google Scholar] [CrossRef]
- Matsubayashi, Y.; Ogawa, M.; Kihara, H.; Niwa, M.; Sakagami, Y. Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 2006, 142, 45–53. [Google Scholar] [CrossRef]
- Ruffel, S.; Krouk, G.; Ristova, D.; Shasha, D.; Birnbaum, K.D.; Coruzzi, G.M. Nitrogen economics of root foraging: Transitive closure of the nitrate–cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc. Natl. Acad. Sci. USA 2011, 108, 18524–18529. [Google Scholar] [CrossRef]
- de Bang, T.C.; Lay, K.S.; Scheible, W.-R.; Takahashi, H. Small peptide signaling pathways modulating macronutrient utilization in plants. Curr. Opin. Plant Biol. 2017, 39, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-L.; Lee, C.-Y.; Cheng, K.-T.; Chang, W.-H.; Huang, R.-N.; Nam, H.G.; Chen, Y.-R. Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 2014, 26, 4135–4148. [Google Scholar] [CrossRef] [PubMed]
- Chien, P.-S.; Nam, H.G.; Chen, Y.-R. A salt-regulated peptide derived from the CAP superfamily protein negatively regulates salt-stress tolerance in Arabidopsis. J. Exp. Bot. 2015, 66, 5301–5313. [Google Scholar] [CrossRef] [PubMed]
- Kellermeier, F.; Armengaud, P.; Seditas, T.J.; Danku, J.; Salt, D.E.; Amtmann, A. Analysis of the root system architecture of arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 2014, 26, 1480–1496. [Google Scholar] [CrossRef] [PubMed]
- Patterson, K.; Cakmak, T.; Cooper, A.; Lager, I.; Rasmusson, A.G.; Escobar, M.A. Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ. 2010, 33, 1486–1501. [Google Scholar] [CrossRef] [PubMed]
- Czyzewicz, N.; Shi, C.-L.; Vu, L.D.; Van De Cotte, B.; Hodgman, C.; Butenko, M.A.; Smet, I.D. Modulation of Arabidopsis and monocot root architecture by CLAVATA3/EMBRYO SURROUNDING REGION 26 peptide. J. Exp. Bot. 2015, 66, 5229–5243. [Google Scholar] [CrossRef] [PubMed]
- Funayama-Noguchi, S.; Noguchi, K.; Yoshida, C.; Kawaguchi, M. Two CLE genes are induced by phosphate in roots of Lotus japonicus. J. Plant Res. 2011, 124, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Lease, K.A.; Walker, J.C. The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 2006, 142, 831–838. [Google Scholar] [CrossRef]
- Ghorbani, S.; Lin, Y.-C.; Parizot, B.; Fernandez, A.; Njo, M.F.; Van de Peer, Y.; Beeckman, T.; Hilson, P. Expanding the repertoire of secretory peptides controlling root development with comparative genome analysis and functional assays. J. Exp. Bot. 2015, 66, 5257–5269. [Google Scholar] [CrossRef]
- Czyzewicz, N.; Wildhagen, M.; Cattaneo, P.; Stahl, Y.; Pinto, K.G.; Aalen, R.B.; Butenko, M.A.; Simon, R.; Hardtke, C.S.; De Smet, I. Antagonistic peptide technology for functional dissection of CLE peptides revisited. J. Exp. Bot. 2015, 66, 5367–5374. [Google Scholar] [CrossRef]
- Strabala, T.J.; O’Donnell, P.J.; Smit, A.-M.; Ampomah-Dwamena, C.; Martin, E.J.; Netzler, N.; Nieuwenhuizen, N.J.; Quinn, B.D.; Foote, H.C.C.; Hudson, K.R. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 2006, 140, 1331–1344. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.L.; Ishida, T.; Yoshimura, M.; Imamura, Y.; Shimaoka, C.; Sawa, S. A collection of mutants for CLE-peptide-encoding genes in Arabidopsis generated by CRISPR/Cas9-mediated gene targeting. Plant Cell Physiol. 2017, 58, 1848–1856. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.A.; Haak, D.C.; Nishimura, M.T.; Teixeira, P.J.P.L.; James, S.R.; Dangl, J.L.; Nimchuk, Z.L. Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PLoS ONE 2016, 11, e0162169. [Google Scholar] [CrossRef] [PubMed]
- Bollig, K.; Lamshöft, M.; Schweimer, K.; Marner, F.-J.; Budzikiewicz, H.; Waffenschmidt, S. Structural analysis of linear hydroxyproline-bound O-glycans of Chlamydomonas reinhardtii—Conservation of the inner core in Chlamydomonas and land plants. Carbohydr. Res. 2007, 342, 2557–2566. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, K.; Shinohara, H.; Ogawa-Ohnishi, M.; Matsubayashi, Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 2009, 5, 578–580. [Google Scholar] [CrossRef] [PubMed]
- DeYoung, B.J.; Clark, S.E. BAM receptors regulate stem cell specification and organ development through complex interactions with CLAVATA signaling. Genetics 2008, 180, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Bleckmann, A.; Weidtkamp-Peters, S.; Seidel, C.A.M.; Simon, R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 2010, 152, 166–176. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, Y.; Li, R.; Song, X.; Wang, Q.; Huang, S.; Jin, J.B.; Liu, C.-M.; Lin, J. Analysis of interactions among the CLAVATA3 receptors reveals a direct interaction between CLAVATA2 and CORYNE in Arabidopsis. Plant J. 2010, 61, 223–233. [Google Scholar] [CrossRef]
- Kinoshita, A.; Betsuyaku, S.; Osakabe, Y.; Mizuno, S.; Nagawa, S.; Stahl, Y.; Simon, R.; Yamaguchi-Shinozaki, K.; Fukuda, H.; Sawa, S. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 2010, 137, 3911–3920. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Han, L.; Hymes, M.; Denver, R.; Clark, S.E. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010, 63, 889–900. [Google Scholar] [CrossRef]
- Stahl, Y.; Grabowski, S.; Bleckmann, A.; Kühnemuth, R.; Weidtkamp-Peters, S.; Pinto, K.G.; Kirschner, G.K.; Schmid, J.B.; Wink, R.H.; Hülsewede, A.; et al. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 2013, 23, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Hazak, O.; Hardtke, C.S. CLAVATA 1-type receptors in plant development. J. Exp. Bot. 2016, 67, 4827–4833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandt, B.; Hothorn, M. SERK co-receptor kinases. Curr. Biol. 2016, 26, R225–R226. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.; Henzler, C.; Hothorn, M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 2013, 341, 889–892. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lay, K.S.; Takahashi, H. Nutrient-Responsive Small Signaling Peptides and Their Influence on the Root System Architecture. Int. J. Mol. Sci. 2018, 19, 3927. https://doi.org/10.3390/ijms19123927
Lay KS, Takahashi H. Nutrient-Responsive Small Signaling Peptides and Their Influence on the Root System Architecture. International Journal of Molecular Sciences. 2018; 19(12):3927. https://doi.org/10.3390/ijms19123927
Chicago/Turabian StyleLay, Katerina S., and Hideki Takahashi. 2018. "Nutrient-Responsive Small Signaling Peptides and Their Influence on the Root System Architecture" International Journal of Molecular Sciences 19, no. 12: 3927. https://doi.org/10.3390/ijms19123927
APA StyleLay, K. S., & Takahashi, H. (2018). Nutrient-Responsive Small Signaling Peptides and Their Influence on the Root System Architecture. International Journal of Molecular Sciences, 19(12), 3927. https://doi.org/10.3390/ijms19123927