Differential Regulation of Thermodynamic Binding Forces of Levocetirizine and (S)-Cetirizine by Lys191 in Human Histamine H1 Receptors
Abstract
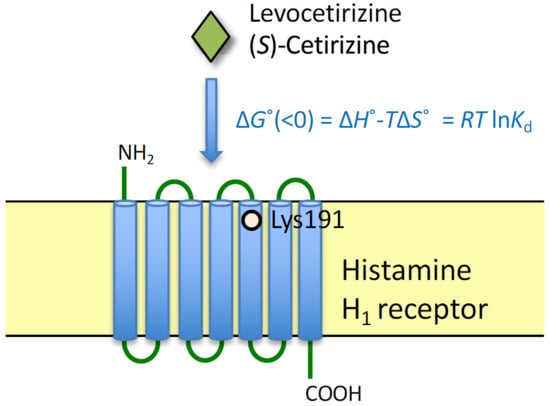
:1. Introduction
2. Results and Discussion
2.1. Binding Affinities of Levocetirizine and (S)-Cetirizine for Hemagglutinin (HA)-Tagged Wild-Type H1 Receptor (HA-WT) and HA-Tagged-Lys191 Mutant of H1 Receptor to Alanine (HA-K191A)
2.2. Thermodynamic Binding Forces of Levocetirizine and (S)-Cetirizine to HA-WT and HA-K191A
3. Materials and Methods
3.1. Materials
3.2. Preparation of Cells and Membrane Fraction
3.3. Measurement of [3H]mepyramine Binding to Membrane Preparations
3.4. Data Analyses
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CHO | Chinese hamster ovary |
| GPCR | G protein-coupled receptor |
| HA | Hemagglutinin |
| K191A | Lys191 mutant of H1 receptor to alanine |
| WT | Wild-type H1 receptor |
References
- Hill, S.J. Distribution, properties, and functional characteristics of three classes of histamine receptor. Pharmacol. Rev. 1990, 42, 45–83. [Google Scholar] [PubMed]
- Hill, S.J.; Ganellin, C.R.; Timmerman, H.; Schwartz, J.C.; Shankley, N.P.; Young, J.M.; Schunack, W.; Levi, R.; Haas, H.L. International Union of Pharmacology. XIII. Classification of histamine receptors. Pharmacol. Rev. 1997, 49, 253–278. [Google Scholar] [PubMed]
- Bakker, R.A.; Timmerman, H.; Leurs, R. Histamine receptors: specific ligands, receptor biochemistry, and signal transduction. Clin. Allergy Immunol. 2002, 17, 27–64. [Google Scholar]
- Yanai, K.; Tashiro, M. The physiological and pathophysiological roles of neuronal histamine: an insight from human positron emission tomography studies. Pharmacol. Ther. 2007, 113, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Haas, H.L.; Sergeeva, O.A.; Selbach, O. Histamine in the nervous system. Physiol. Rev. 2008, 88, 1183–1241. [Google Scholar] [CrossRef]
- Holgate, S.T.; Canonica, G.W.; Simons, F.E.; Taglialatela, M.; Tharp, M.; Timmerman, H.; Yanai, K. Consensus group on new-generation antihistamines (CONGA): present status and recommendations. Clin. Exp. Allergy 2003, 33, 1305–1324. [Google Scholar] [CrossRef] [PubMed]
- Kalpaklioglu, F.; Baccioglu, A. Efficacy and safety of H1-antihistamines: an update. Antiinflamm. Antiallergy Agents Med. Chem. 2012, 11, 230–237. [Google Scholar] [CrossRef]
- Yanai, K.; Yoshikawa, T.; Yanai, A.; Nakamura, T.; Iida, T.; Leurs, R.; Tashiro, M. The clinical pharmacology of non-sedating antihistamines. Pharmacol. Ther. 2017, 178, 148–156. [Google Scholar] [CrossRef]
- Church, M.K. Allergy, histamine and antihistamines. Handb. Exp. Pharmacol. 2017, 241, 321–331. [Google Scholar]
- Tillement, J.P.; Testa, B.; Brée, F. Compared pharmacological characteristics in humans of racemic cetirizine and levocetirizine, two histamine H1-receptor antagonists. Biochem. Pharmacol. 2003, 66, 1123–1126. [Google Scholar] [CrossRef]
- Chen, C. Physicochemical, pharmacological and pharmacokinetic properties of the zwitterionic antihistamines cetirizine and levocetirizine. Curr. Med. Chem. 2008, 15, 2173–2191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cheng, L.; Hong, J. The clinical use of cetirizine in the treatment of allergic rhinitis. Pharmacology 2013, 92, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Gillard, M.; Van der Perren, C.; Moguilevsky, N.; Massingham, R.; Chatelain, P. Binding characteristics of cetirizine and levocetirizine to human H1 histamine receptors: contribution of Lys191 and Thr194. Mol. Pharmacol. 2002, 61, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Shimamura, T.; Shiroishi, M.; Weyand, S.; Tsujimoto, H.; Winter, G.; Katritch, V.; Abagyan, R.; Cherezov, V.; Liu, W.; Han, G.W.; et al. Structure of the human histamine H1 receptor complex with doxepin. Nature 2011, 475, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Hitzemann, R. Thermodynamics aspects of drug–receptor interactions. Trends Pharm. Sci. 1988, 9, 408–411. [Google Scholar] [CrossRef]
- Borea, P.A.; Dalpiaz, A.; Varani, K.; Gilli, P.; Gilli, G. Can thermodynamic measurements of receptor binding yield information on drug affinity and efficacy? Biochem. Pharmacol. 2000, 60, 1549–1556. [Google Scholar] [CrossRef]
- Wittmann, H.J.; Seifert, R.; Strasser, A. Contribution of binding enthalpy and entropy to affinity of antagonist and agonist binding at human and guinea pig histamine H1-receptor. Mol. Pharmacol. 2009, 76, 25–37. [Google Scholar] [CrossRef]
- Hishinuma, S.; Sugawara, K.; Uesawa, Y.; Fukui, H.; Shoji, M. Differential thermodynamic driving force of first- and second-generation antihistamines to determine their binding affinity for human H1 receptors. Biochem. Pharmacol. 2014, 91, 231–241. [Google Scholar] [CrossRef]
- Hishinuma, S.; Komazaki, H.; Tsukamoto, H.; Hatahara, H.; Fukui, H.; Shoji, M. Ca2+-dependent down-regulation of human histamine H1 receptors in Chinese hamster ovary cells. J. Neurochem. 2018, 144, 68–80. [Google Scholar] [CrossRef]
- Hishinuma, S.; Nozawa, H.; Akatsu, C.; Shoji, M. C-terminal of human histamine H1 receptors regulates their agonist-induced clathrin-mediated internalization and G-protein signaling. J. Neurochem. 2016, 139, 552–565. [Google Scholar] [CrossRef]
- Hishinuma, S.; Kosaka, K.; Akatsu, C.; Uesawa, Y.; Fukui, H.; Shoji, M. Asp73-dependent and -independent regulation of the affinity of ligands for human histamine H1 receptors by Na+. Biochem. Pharmacol. 2017, 128, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Prusoff, W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar]




| Antihistamines | Binding Affinity | Thermodynamic Binding Forces | |
|---|---|---|---|
| Enthalpy-Dependent Electrostatic Binding Forces | Entropy-Dependent Hydrophobic Binding Forces | ||
| Levocetirizine | ↓** | ↑* | ↓* |
| (S)-Cetirizine | ↓** | ↓** | ↑** |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hishinuma, S.; Tamura, Y.; Kobayashi, C.; Akatsu, C.; Shoji, M. Differential Regulation of Thermodynamic Binding Forces of Levocetirizine and (S)-Cetirizine by Lys191 in Human Histamine H1 Receptors. Int. J. Mol. Sci. 2018, 19, 4067. https://doi.org/10.3390/ijms19124067
Hishinuma S, Tamura Y, Kobayashi C, Akatsu C, Shoji M. Differential Regulation of Thermodynamic Binding Forces of Levocetirizine and (S)-Cetirizine by Lys191 in Human Histamine H1 Receptors. International Journal of Molecular Sciences. 2018; 19(12):4067. https://doi.org/10.3390/ijms19124067
Chicago/Turabian StyleHishinuma, Shigeru, Yuri Tamura, Chihiro Kobayashi, Chizuru Akatsu, and Masaru Shoji. 2018. "Differential Regulation of Thermodynamic Binding Forces of Levocetirizine and (S)-Cetirizine by Lys191 in Human Histamine H1 Receptors" International Journal of Molecular Sciences 19, no. 12: 4067. https://doi.org/10.3390/ijms19124067
APA StyleHishinuma, S., Tamura, Y., Kobayashi, C., Akatsu, C., & Shoji, M. (2018). Differential Regulation of Thermodynamic Binding Forces of Levocetirizine and (S)-Cetirizine by Lys191 in Human Histamine H1 Receptors. International Journal of Molecular Sciences, 19(12), 4067. https://doi.org/10.3390/ijms19124067