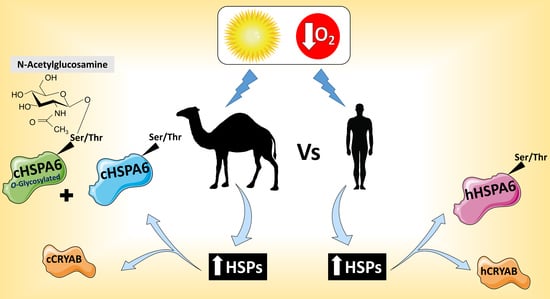
Differential Glycosylation and Modulation of Camel and Human HSP Isoforms in Response to Thermal and Hypoxic Stresses
Abstract
:1. Introduction
2. Results
2.1. Cloning, Expression and Localization of Camel and Human HSPs in COS-1 Cells
2.2. Glycosylation of Camel and Human HSPs
2.3. Response of Camel and Human HSPA6 Isoforms to Heat and Low Oxygen Stress Conditions
3. Discussion
4. Materials and Methods
4.1. Immunological Reagents
4.2. Sample Collection, RNA Isolation, cDNA Synthesis and Subcloning
4.3. Transient Transfection of COS-1 Cells, Western Blotting, Biosynthetic Labeling, and Immunoprecipitation
4.4. Indirect Immunofluorescence Analysis
4.5. Characterization of the Protein Glycosylation in Camel and Human HSPA6
4.6. Treatment with Different Temperatures and Low Oxygen Conditions
4.7. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| HSP | Heat shock protein |
| O-GlcNAc | O-linked β-N-acetylglucosamine |
| PTM | Post-translational modification |
Appendix A. The cDNA and Deduced Protein Sequence of Camel HSPA6

Appendix B. The Expression and Mass Spectrometry Analysis of Camel and Human HSPA6

| Protein Band | Protein | Score | Protein Mass (kDa) | Unique Peptide Number | Coverage % |
|---|---|---|---|---|---|
| a | cHSPA6 | 10,069 | 75.8 | 30 | 86.44 |
| b | cHSPA6 | 2416.03 | 75.8 | 19 | 65.31 |
| c | hHSPA6 | 2112.6 | 76.2 | 19 | 66.62 |
| d | hHSPA6 | 4407 | 76.2 | 23 | 71.87 |
| e | hHSPA6 | 6215.7 | 76.2 | 29 | 73.62 |
Appendix C. Binding of cHSPA6 and hHSPA6 to Helix pomatia Lectin under Normal Culture Conditions and Stress Conditions

References
- Schmidt-Nilsen, K. Animals of the Deserts; Nauka: Moscow, Russia, 1972. [Google Scholar]
- Elkhawad, A.O. Selective brain cooling in desert animals: The camel (Camelus dromedarius). Comp. Biochem. Physiol. Part A Physiol. 1992, 101, 195–201. [Google Scholar] [CrossRef]
- Ouajd, S.; Kamel, B. Physiological particularities of dromedary (Camelus dromedarius) and experimental implications. Scand. J. Lab. Anim. Sci. 2009, 36, 19–29. [Google Scholar]
- Thayyullathil, F.; Chathoth, S.; Hago, A.; Wernery, U.; Patel, M.; Galadari, S. Investigation of heat stress response in the camel fibroblast cell line dubca. Ann. N. Y. Acad. Sci. 2008, 1138, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, H.A.; Karaev, K.K.; Lyashko, V.N.; Evgen’ev, M.B. Heat-shock response in camel (Camelus dromedarius) blood cells and adaptation to hyperthermia. Comp. Biochem. Physiol. B 1993, 106, 867–872. [Google Scholar] [CrossRef]
- Jackson, S.E. Hsp90: Structure and function. Top. Curr. Chem. 2013, 328, 155–240. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.; Jäger, R.; Mosser, D.D.; Samali, A. Regulation of apoptosis by heat shock proteins. IUBMB Life 2014, 66, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Helmbrecht, K.; Zeise, E.; Rensing, L. Chaperones in cell cycle regulation and mitogenic signal transduction: A review. Cell Prolif. 2000, 33, 341–365. [Google Scholar] [CrossRef] [PubMed]
- Haslbeck, M. sHsps and their role in the chaperone network. Cell. Mol. Life Sci. 2002, 59, 1649–1657. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, J.-M.; Sun, X.; Benndorf, R.; Welsh, M.J. Interactions of HSP22 (HSPB8) with HSP20, alphaB-crystallin, and HSPB3. Biochem. Biophys. Res. Commun. 2005, 337, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Vos, M.J.; Hageman, J.; Carra, S.; Kampinga, H.H. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry 2008, 47, 7001–7011. [Google Scholar] [CrossRef] [PubMed]
- Heads, R.J.; Yellon, D.M.; Latchman, D.S. Differential cytoprotection against heat stress or hypoxia following expression of specific stress protein genes in myogenic cells. J. Mol. Cell. Cardiol. 1995, 27, 1669–1678. [Google Scholar] [CrossRef]
- Mymrikov, E.V.; Seit-Nebi, A.S.; Gusev, N.B. Large potentials of small heat shock proteins. Physiol. Rev. 2011, 91, 1123–1159. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, J. Alpha-crystallin can function as a molecular chaperone. Proc. Natl. Acad. Sci. USA 1992, 89, 10449–10453. [Google Scholar] [CrossRef] [PubMed]
- Carver, J.A.; Aquilina, J.A.; Cooper, P.G.; Williams, G.A.; Truscott, R.J. Alpha-crystallin: Molecular chaperone and protein surfactant. Biochim. Biophys. Acta 1994, 1204, 195–206. [Google Scholar] [CrossRef]
- Tang, S.; Yin, B.; Song, E.; Chen, H.; Cheng, Y.; Zhang, X.; Bao, E.; Hartung, J. Aspirin upregulates αB-Crystallin to protect the myocardium against heat stress in broiler chickens. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Lv, Y.; Chen, H.; Adam, A.; Cheng, Y.; Hartung, J.; Bao, E. Comparative analysis of alpha-B-crystallin expression in heat-stressed myocardial cells in vivo and in vitro. PLoS ONE 2014, 9, e86937. [Google Scholar]
- Mchaourab, H.S.; Godar, J.A.; Stewart, P.L. Structure and mechanism of protein stability sensors: Chaperone activity of small heat shock proteins. Biochemistry 2009, 48, 3828–3837. [Google Scholar] [CrossRef] [PubMed]
- Bakthisaran, R.; Akula, K.K.; Tangirala, R.; Rao, C.M. Ohan phosphorylation of αB-crystallin: Role in stress, aging and patho-physiological conditions. Biochim. Biophys. Acta 2016, 1860, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Van de Schootbrugge, C.; Schults, E.M.J.; Bussink, J.; Span, P.N.; Grénman, R.; Pruijn, G.J.M.; Kaanders, J.H.; Boelens, W.C. Effect of hypoxia on the expression of αB-crystallin in head and neck squamous cell carcinoma. BMC Cancer 2014, 14, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van De Klundert, F.A.J.M.; De Jong, W.W. The small heat shock proteins Hsp20 and αB-crystallin in cultured cardiac myocytes: Differences in cellular localization and solubilization after heat stress. Eur. J. Cell Biol. 1999, 78, 567–572. [Google Scholar] [CrossRef]
- Kusukawa, N.; Yura, T. Heat shock protein GroE of Escherichia coli: Key protective roles against thermal stress. Genes Dev. 1988, 2, 874–882. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhao, T.; Huang, X.; Liu, Z.H.; Xiong, L.; Li, M.M.; Wu, L.Y.; Zhao, Y.Q.; Zhu, L.L.; Fan, M. Preinduction of HSP70 promotes hypoxic tolerance and facilitates acclimatization to acute hypobaric hypoxia in mouse brain. Cell Stress Chaperones 2009, 14, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Noonan, E.J.; Place, R.F.; Giardina, C.; Hightower, L.E. Hsp70B’ regulation and function. Cell Stress Chaperones 2007, 12, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Wada, K.-I.; Taniguchi, A.; Xu, L.; Okano, T. Rapid and highly sensitive detection of cadmium chloride induced cytotoxicity using the HSP70B’ promoter in live cells. Biotechnol. Bioeng. 2005, 92, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Leung, T.K.; Rajendran, M.Y.; Monfries, C.; Hall, C.; Lim, L. The human heat-shock protein family. Expression of a novel heat-inducible HSP70 (HSP70B’) and isolation of its cDNA and genomic DNA. Biochem. J. 1990, 267, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Parsian, A.J.; Sheren, J.E.; Tao, T.Y.; Goswami, P.C.; Malyapa, R.; Van Rheeden, R.; Watson, M.S.; Hunt, C.R. The human Hsp70B gene at the HSPA7 locus of chromosome 1 is transcribed but non-functional. Biochim. Biophys. Acta 2000, 1494, 201–205. [Google Scholar] [CrossRef]
- Hageman, J.; van Waarde, M.A.W.H.; Zylicz, A.; Walerych, D.; Kampinga, H.H. The diverse members of the mammalian HSP70 machine show distinct chaperone-like activities. Biochem. J. 2011, 435, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Daugaard, M.; Rohde, M.; Jäättelä, M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007, 581, 3702–3710. [Google Scholar] [CrossRef] [PubMed]
- Noonan, E.J.; Fournier, G.; Hightower, L.E. Surface expression of HSP70B’ in response to proteasome inhibition in human colon cells. Cell Stress Chaperones 2008, 13, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Khalouei, S.; Chow, A.M.; Brown, I.R. Localization of heat shock protein HSPA6 (HSP70B’) to sites of transcription in cultured differentiated human neuronal cells following thermal stress. J. Neurochem. 2014, 131, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhuang, L.; Szatmary, P.; Wen, L.; Sun, H.; Lu, Y.; Xu, Q.; Chen, X. Upregulation of heat shock proteins (HSPA12A, HSP90B1, HSPA4, HSPA5 and HSPA6) in tumour tissues is associated with poor outcomes from HBV-related early-stage hepatocellular carcinoma. Int. J. Med. Sci. 2015, 12, 256–263. [Google Scholar] [CrossRef] [PubMed]
- McCullagh, K.J.A.; Cooney, R.; O’Brien, T. Endothelial nitric oxide synthase induces heat shock protein HSPA6 (HSP70B’) in human arterial smooth muscle cells. Nitric Oxide 2016, 52, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Sainis, I.; Angelidis, C.; Pagoulatos, G.; Lazaridis, I. The HSC70 gene which is slightly induced by heat is the main virus inducible member of the HSP70 gene family. FEBS Lett. 1994, 355, 282–286. [Google Scholar] [CrossRef]
- Pirillo, A.; Norata, G.D.; Zanelli, T.; Catapano, A.L. Overexpression of inducible heat shock protein 70 in COS-1 cells fails to protect from cytotoxicity of oxidized LDLS. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Zachara, N.E.; Hart, G.W. O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta 2004, 1673, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Zachara, N.E.; Donnell, N.O.; Cheung, W.D.; Mercer, J.J.; Marth, J.D.; Hart, G.W. Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to Stress—A survival response of mammalian cells. J. Biol. Chem. 2004, 279, 30133–30142. [Google Scholar] [CrossRef] [PubMed]
- Rambaruth, N.D.; Greenwell, P.; Dwek, M.V. The lectin Helix pomatia agglutinin recognizes O-GlcNAc containing glycoproteins in human breast cancer. Glycobiology 2012, 22, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Sonna, L.A.; Cullivan, M.L.; Sheldon, H.K.; Pratt, R.E.; Lilly, C.M. Effect of hypoxia on gene expression by human hepatocytes (HepG2). Physiol. Genom. 2003, 12, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.A.; Goping, I.S.; Berry, F.; Walter, M.A. Dysfunction of the stress-responsive FOXC1 transcription factor contributes to the earlier-onset glaucoma observed in Axenfeld-Rieger syndrome patients. Cell Death Dis. 2014, 5, e1069. [Google Scholar] [CrossRef] [PubMed]
- Louapre, P.; Grongnet, J.F.; Tanguay, R.M.; David, J.C. Effects of hypoxia on stress proteins in the piglet heart at birth. Cell Stress Chaperones 2005, 10, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Mer, G.; Hietter, H.; Lefèvre, J.F. Stabilization of proteins by glycosylation examined by NMR analysis of a fucosylated proteinase inhibitor. Nat. Struct. Biol. 1996, 3, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, T.; Cieniewski, C.; Lemoine, J.; Guerardel, Y.; Leroy, Y.; Zanetta, J.P.; Michalski, J.C. Identification of N-acetyl-d-glucosamine-specific lectins from rat liver cytosolic and nuclear compartments as heat-shock proteins. Biochem. J. 2001, 360, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Moley, K.H.; Mueckler, M.M. Glucose transport and apoptosis. Apoptosis 2000, 5, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Wells, L.; Vosseller, K.; Cole, R.N.; Cronshaw, J.M.; Matunis, M.J.; Hart, G.W. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteom. 2002, 1, 791–804. [Google Scholar] [CrossRef]
- Walgren, J.L.E.; Vincent, T.S.; Schey, K.L.; Buse, M.G. High glucose and insulin promote O-GlcNAc modification of proteins, including alpha-tubulin. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E424–E434. [Google Scholar] [CrossRef] [PubMed]
- Roquemore, E.P.; Chevrier, M.R.; Cotter, R.J.; Hart, G.W. Dynamic O-GlcNAcylation of the small heat shock protein alpha B-crystallin. Biochemistry 1996, 35, 3578–3586. [Google Scholar] [CrossRef] [PubMed]
- Roquemore, E.P.; Dell, A.; Morris, H.R.; Panico, M.; Reason, A.J.; Savoy, L.A.; Wistow, G.J.; Zigler, J.S.; Earles, B.J.; Hart, G.W. Vertebrate lens α-crystallins are modified by O-linked N-acetylglucosamine. J. Biol. Chem. 1992, 267, 555–563. [Google Scholar] [PubMed]
- Basha, E.; O’Neill, H.; Vierling, E. Small heat shock proteins and α-crystallins: Dynamic proteins with flexible functions. Trends Biochem. Sci. 2012, 37, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Shinohara, H.; Goto, S.; Inaguma, Y.; Morishita, R.; Asano, T. Copurification of small heat shock protein with α B crystallin from human skeletal muscle. J. Biol. Chem. 1992, 267, 7718–7725. [Google Scholar] [PubMed]
- Chow, A.M.; Mok, P.; Xiao, D.; Khalouei, S.; Brown, I.R. Heteromeric complexes of heat shock protein 70 (HSP70) family members, including Hsp70B′, in differentiated human neuronal cells. Cell Stress Chaperones 2010, 15, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Jagan Mohanarao, G.; Mukherjee, A.; Banerjee, D.; Gohain, M.; Dass, G.; Brahma, B.; Datta, T.K.; Upadhyay, R.C.; De, S. HSP70 family genes and HSP27 expression in response to heat and cold stress in vitro in peripheral blood mononuclear cells of goat (Capra hircus). Small Rumin. Res. 2014, 116, 94–99. [Google Scholar] [CrossRef]
- Khalouei, S.; Chow, A.M.; Brown, I.R. Stress-induced localization of HSPA6 (HSP70B’) and HSPA1A (HSP70-1) proteins to centrioles in human neuronal cells. Cell Stress Chaperones 2014, 19, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Sadder, M.T.; Migdadi, H.M.; Zakri, A.M.; Abdoun, K.A.; Samara, E.M.; Okab, A.B.; Al-Haidary, A.A. Expression analysis of heat shock proteins in dromedary camel (Camelus dromedarius). J. Camel Pract. Res. 2015, 22, 19. [Google Scholar] [CrossRef]
- Du, X.L.; Edelstein, D.; Dimmeler, S.; Ju, Q.; Sui, C.; Brownlee, M. Hyperglycemia inhibits endothelial nitric oxide synthase activity by posttranslational modification at the Akt site. J. Clin. Investig. 2001, 108, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Federici, M. Insulin-dependent activation of endothelial nitric oxide synthase is impaired by O-linked glycosylation modification of signaling proteins in human coronary endothelial cells. Circulation 2002, 106, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Parker, G.J.; Lund, K.C.; Taylor, R.P.; McClain, D.A. Insulin resistance of glycogen synthase mediated by O-linked N-acetylglucosamine. J. Biol. Chem. 2003, 278, 10022–10027. [Google Scholar] [CrossRef] [PubMed]
- Han, I.; Roos, M.D.; Kudlow, J.E. Interaction of the transcription factor Sp1 with the nuclear pore protein p62 requires the C-terminal domain of p62. J. Cell. Biochem. 1998, 68, 50–61. [Google Scholar] [CrossRef]
- Roos, M.D.; Su, K.; Baker, J.R.; Kudlow, J.E. O glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol. Cell. Biol. 1997, 17, 6472–6780. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Cole, R.N.; Zaia, J.; Hart, G.W. Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor beta. Biochemistry 2000, 39, 11609–11620. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Miyazaki, J.I.; Hart, G.W. The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 β-cells. Arch. Biochem. Biophys. 2003, 415, 155–163. [Google Scholar] [CrossRef]
- Naim, H.Y.; Lacey, S.W.; Sambrook, J.F.; Gething, M.J.H. Expression of a full-length cDNA coding for human intestinal lactase-phlorizin hydrolase reveals an uncleaved, enzymatically active, and transport-competent protein. J. Biol. Chem. 1991, 266, 12313–12320. [Google Scholar] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.; Weiner, J.R.; Stadge, S.; Naim, H.Y. Additional N-glycosylation and its impact on the folding of intestinal lactase-phlorizin hydrolase. J. Biol. Chem. 2000, 275, 10630–10637. [Google Scholar] [CrossRef] [PubMed]
- Dyballa, N.; Metzger, S. Fast and sensitive colloidal coomassie G-250 staining for proteins in polyacrylamide gels. J. Vis. Exp. 2009, 2–5. [Google Scholar] [CrossRef] [PubMed]






| Residue Number | Residue | O-Glc-NAc Result 1 | Potential | Threshold 1 | Threshold 2 |
|---|---|---|---|---|---|
| Camel CRYAB | |||||
| 41 | T | +++ | 0.6512 | 0.3982 | 0.4871 |
| 42 | T | + | 0.4726 | 0.3966 | 0.4850 |
| 43 | S | ++ | 0.5398 | 0.4026 | 0.4931 |
| 53 | S | + | 0.4490 | 0.3919 | 0.4786 |
| 134 | T | ++ | 0.6134 | 0.4376 | 0.5402 |
| 135 | S | ++ | 0.5522 | 0.4257 | 0.5242 |
| 153 | S | + | 0.3793 | 0.3276 | 0.3919 |
| 170 | T | +++ | 0.6383 | 0.3557 | 0.4299 |
| Human CRYAB | |||||
| 41 | S | +++ | 0.6443 | 0.3921 | 0.4790 |
| 42 | T | + | 0.4260 | 0.3923 | 0.4792 |
| 43 | S | ++ | 0.4972 | 0.3990 | 0.4882 |
| 53 | S | + | 0.4520 | 0.3907 | 0.4770 |
| 132 | T | ++ | 0.5897 | 0.4157 | 0.5107 |
| 134 | T | ++ | 0.5798 | 0.4332 | 0.5343 |
| 135 | S | ++ | 0.5551 | 0.4190 | 0.5152 |
| 153 | S | +++ | 0.5567 | 0.3481 | 0.4196 |
| 170 | T | +++ | 0.6383 | 0.3557 | 0.4299 |
| Camel HSPA6 | |||||
| 40 | T | + | 0.5184 | 0.4261 | 0.5247 |
| 160 | T | + | 0.4538 | 0.3931 | 0.4803 |
| 275 | T | + | 0.442 | 0.4185 | 0.5145 |
| 427 | T | + | 0.4854 | 0.424 | 0.5219 |
| 432 | T | + | 0.5234 | 0.4549 | 0.5636 |
| 434 | S | + | 0.4552 | 0.4344 | 0.5359 |
| 621 | S | ++ | 0.5823 | 0.4446 | 0.5497 |
| 634 | S | + | 0.4399 | 0.41 | 0.5031 |
| 635 | T | + | 0.4453 | 0.4263 | 0.525 |
| Human HSPA6 | |||||
| 40 | T | + | 0.5184 | 0.4261 | 0.5247 |
| 160 | T | + | 0.4538 | 0.3931 | 0.4803 |
| 275 | T | + | 0.442 | 0.4185 | 0.5145 |
| 427 | T | + | 0.4854 | 0.424 | 0.5219 |
| 432 | T | + | 0.5056 | 0.4518 | 0.5594 |
| Target cDNA | (Protein_ID) GenBank | Camel HSPs Primers |
| CRYAB | AHL21625.1 | Forward 5′-ATGGACATCGCCATCCACC-3′ Reverse 5′-CTATTTCTTGGGGGCTGCGG-3′ |
| HSPA6 | ADO12067.1 | Forward 5′-ATGTCTGCCGCAAAGGAAGTGG-3′ Reverse 5′-TTAATCAACCTCCTCAATAACAGGGCCA-3′ |
| Human HSPs Primers | ||
| CRYAB | AAP35416.1 | Forward 5′-ATGGACATCGCCATCCACC-3′ Reverse 5′-CTATTTCTTGGGGGCTGCGG-3′ |
| HSPA6 | NP_002146.2 | Forward 5′-ATGCAGGCCCCACGGGAG-3′ Reverse 5′-TCAATCAACCTCCTCAATGATGGGGCC-3′ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoter, A.; Amiri, M.; Prince, A.; Amer, H.; Warda, M.; Naim, H.Y. Differential Glycosylation and Modulation of Camel and Human HSP Isoforms in Response to Thermal and Hypoxic Stresses. Int. J. Mol. Sci. 2018, 19, 402. https://doi.org/10.3390/ijms19020402
Hoter A, Amiri M, Prince A, Amer H, Warda M, Naim HY. Differential Glycosylation and Modulation of Camel and Human HSP Isoforms in Response to Thermal and Hypoxic Stresses. International Journal of Molecular Sciences. 2018; 19(2):402. https://doi.org/10.3390/ijms19020402
Chicago/Turabian StyleHoter, Abdullah, Mahdi Amiri, Abdelbary Prince, Hassan Amer, Mohamad Warda, and Hassan Y. Naim. 2018. "Differential Glycosylation and Modulation of Camel and Human HSP Isoforms in Response to Thermal and Hypoxic Stresses" International Journal of Molecular Sciences 19, no. 2: 402. https://doi.org/10.3390/ijms19020402
APA StyleHoter, A., Amiri, M., Prince, A., Amer, H., Warda, M., & Naim, H. Y. (2018). Differential Glycosylation and Modulation of Camel and Human HSP Isoforms in Response to Thermal and Hypoxic Stresses. International Journal of Molecular Sciences, 19(2), 402. https://doi.org/10.3390/ijms19020402