The PUF Protein Family: Overview on PUF RNA Targets, Biological Functions, and Post Transcriptional Regulation
Abstract
:1. Introduction
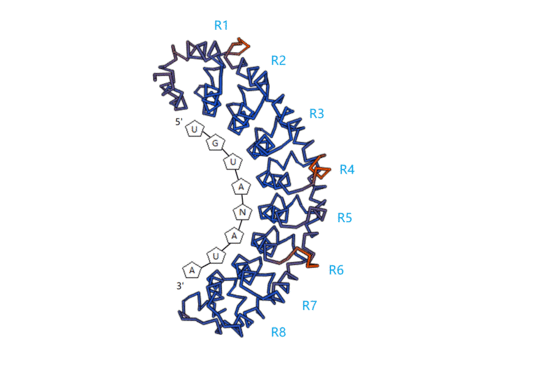
2. RNA-Binding Target of PUF Proteins
3. Putative Biological Functions of PUF Proteins
4. PUF Proteins Control Post-Transcriptional Processes through Different Mechanisms
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Wickens, M.; Bernstein, D.S.; Kimble, J.; Parker, R. A PUF family portrait: 3′ UTR regulation as a way of life. Trends Genet. 2002, 18, 150–157. [Google Scholar] [CrossRef]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008, 582, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007, 8, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, N.; Kim, H.B.; Park, N.I.; Kim, H.S.; Kim, Y.K.; Park, Y.I.; Choi, S.B. APUM23, a nucleolar PUF domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010, 64, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.; Kazan, H.; Cook, K.B.; Weirauch, M.T.; Najafabadi, H.S.; Li, X.; Gueroussov, S.; Albu, M.; Zheng, H.; Yang, A.; et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature 2013, 499, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhu, D.; Lin, X.; Miao, J.; Gu, L.; Deng, X.; Yang, Q.; Sun, K.; Zhu, D.; Cao, X.; et al. RNA-binding proteins At RZ-1B and At RZ-1C play a critical role in regulation of pre-mRNA splicing and gene expression during Arabidopsis development. Plant Cell 2016, 28, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Tam, P.P.; Barrette-Ng, I.H.; Simon, D.M.; Tam, M.W.; Ang, A.L.; Muench, D.G. The PUF family of RNA-binding proteins in plants: Phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 2010, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Friend, K.; Campbell, Z.T.; Cooke, A.; Kroll-Conner, P.; Wickens, M.P.; Kimble, J. A conserved PUF–Ago–eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 2012, 19, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Van Etten, J.; Schagat, T.L.; Hrit, J.; Weidmann, C.A.; Brumbaugh, J.; Coon, J.J.; Goldstrohm, A.C. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J. Biol. Chem. 2012, 287, 36370–36383. [Google Scholar] [CrossRef] [PubMed]
- Miles, W.O.; Tschöp, K.; Herr, A.; Ji, J.Y.; Dyson, N.J. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012, 26, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kopp, F.; Chang, T.C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD regulates genomic stability by sequestering Pumilio proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Deng, Y.; Zenklusen, D.; Singer, R.H. A new yeast PUF family protein, PUF6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004, 18, 1452–1465. [Google Scholar] [CrossRef] [PubMed]
- Suh, N.; Crittenden, S.L.; Goldstrohm, A.; Hook, B.; Thompson, B.; Wickens, M.; Kimble, J. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics 2009, 181, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Gallegos, M.; Puoti, A.; Durkin, E.; Fields, S.; Kimble, J.; Wickens, M.P. A conserved RNA-binding protein that regulates sexual fates in the Caenorhabditis elegans hermaphrodite germ line. Nature 1997, 390, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.D.; Wang, C.; Moore, J.; Dickinson, L.K.; Lehmann, R. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev. 1992, 6, 2312–2326. [Google Scholar] [CrossRef] [PubMed]
- Zamore, P.D.; Williamson, J.R.; Lehmann, R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 1997, 3, 1421–1433. [Google Scholar] [PubMed]
- Wang, X.; Zamore, P.D.; Hall, T.M.T. Crystal structure of a Pumilio homology domain. Mol. Cell 2001, 7, 855–865. [Google Scholar] [CrossRef]
- Hall, T.M.T. De-coding and re-coding RNA recognition by PUF and PPR repeat proteins. Curr. Opin. Struct. Biol. 2016, 36, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; McLachlan, J.; Zamore, P.D.; Hall, T.M.T. Modular recognition of RNA by a human pumilio-homology domain. Cell 2002, 110, 501–512. [Google Scholar] [CrossRef]
- Ahringer, J.; Kimble, J. Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3′ untranslated region. Nature 1991, 349, 346–348. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, J.; Wharton, R.P. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999, 13, 2704–2712. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, S.; Katsu, Y.; Mita, K.; Inoue, K.; Nagahama, Y.; Yamashita, M. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 2001, 276, 20945–20953. [Google Scholar] [CrossRef] [PubMed]
- Tadauchi, T.; Matsumoto, K.; Herskowitz, I.; Irie, K. Post-transcriptional regulation through the HO 3′ UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 2001, 20, 552–561. [Google Scholar] [CrossRef] [PubMed]
- Crittenden, S.L.; Bernstein, D.S.; Bachorik, J.L.; Thompson, B.E.; Gallegos, M.; Petcherski, A.G.; Moulder, G.; Barstead, R.; Wickens, M.; Kimble, J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 2002, 417, 660–663. [Google Scholar] [CrossRef] [PubMed]
- Spassov, D.; Jurecic, R. The PUF Family of RNA-binding Proteins: Does Evolutionarily Conserved Structure Equal Conserved Function? IUBMB Life 2003, 55, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Galgano, A.; Forrer, M.; Jaskiewicz, L.; Kanitz, A.; Zavolan, M.; Gerber, A.P. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS ONE 2008, 3, e3164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamore, P.D.; Bartel, D.P.; Lehmann, R.; Williamson, J.R. The PUMILIO-RNA Interaction: A single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry 1999, 38, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Gupta, Y.K.; Lee, T.H.; Edwards, T.A.; Escalante, C.R.; Kadyrova, L.Y.; Wharton, R.P.; Aggarwal, A.K. Co-occupancy of two Pumilio molecules on a single hunchback NRE. RNA 2009, 15, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, D.; Hook, B.; Hajarnavis, A.; Opperman, L.; Wickens, M. Binding specificity and mRNA targets of a Caenorhabditis elegans PUF protein, FBF-1. RNA 2005, 11, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Kershner, A.; Wang, Y.; Holley, C.P.; Wilinski, D.; Keles, S.; Kimble, J.; Wickens, M.; Hall, T.M. Divergence of Pumilio/fem-3 mRNA binding factor (PUF) protein specificity through variations in an RNA-binding pocket. J. Biol. Chem. 2012, 287, 6949–6957. [Google Scholar] [CrossRef] [PubMed]
- Valley, C.T.; Porter, D.F.; Qiu, C.; Campbell, Z.T.; Hall, T.M.T.; Wickens, M. Patterns and plasticity in RNA-protein interactions enable recruitment of multiple proteins through a single site. Proc. Natl. Acad. Sci. USA 2012, 109, 6054–6059. [Google Scholar] [CrossRef] [PubMed]
- García-Rodríguez, L.J.; Gay, A.C.; Pon, L.A. PUF3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J. Cell Biol. 2007, 176, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.P.; Herschlag, D.; Brown, P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004, 2, e79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porter, D.F.; Koh, Y.Y.; VanVeller, B.; Raines, R.T.; Wickens, M. Target selection by natural and redesigned PUF proteins. Proc. Natl. Acad. Sci. USA 2015, 112, 15868–15873. [Google Scholar] [CrossRef] [PubMed]
- White, E.K.; Moore-Jarrett, T.; Ruley, H.E. PUM2, a novel murine PUF protein, and its consensus RNA-binding site. RNA 2001, 7, 1855–1866. [Google Scholar] [PubMed]
- Lu, G.; Dolgner, S.J.; Hall, T.M.T. Understanding and engineering RNA sequence specificity of PUF proteins. Curr. Opin. Struct. Biol. 2009, 19, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Cheong, C.G.; Hall, T.M.T. Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci. USA 2006, 103, 13635–13639. [Google Scholar] [CrossRef] [PubMed]
- Gupta, Y.K.; Nair, D.T.; Wharton, R.P.; Aggarwal, A.K. Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity. Structure 2008, 16, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.T.; Higgin, J.J.; Hall, T.M.T. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat. Struct. Mol. Biol. 2008, 15, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Muraro, N.I.; Weston, A.J.; Gerber, A.P.; Luschnig, S.; Moffat, K.G.; Baines, R.A. Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J. Neurosci. 2008, 28, 2099–2109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, J.N.; Panepinto, J.C. Morphotype-specific effector functions of Cryptococcus neoformans PUM1. Sci. Rep. 2016, 6, 23638. [Google Scholar] [CrossRef] [PubMed]
- Francischini, C.W.; Quaggio, R.B. Molecular characterization of Arabidopsis thaliana PUF proteins–binding specificity and target candidates. FEBS J. 2009, 276, 5456–5470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Muench, D.G. A nucleolar PUF RNA-binding protein with specificity for a unique RNA sequence. J. Biol. Chem. 2015, 290, 30108–30118. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, F.; Arnold, K.; Künzli, M.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009, 37, D387–D392. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, B.; Crittenden, S.; Gallegos, M.; Moulder, G.; Barstead, R.; Kimble, J.; Wickens, M. NANOS-3 and FBF proteins physically interact to control the sperm–oocyte switch in Caenorhabditis elegans. Curr. Biol. 1999, 9, 1009–1018. [Google Scholar] [CrossRef]
- Stein, L.; Sternberg, P.; Durbin, R.; Thierry-Mieg, J.; Spieth, J. WormBase: Network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 2001, 29, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Stumpf, C.R.; Kimble, J.; Wickens, M. A Caenorhabditis elegans PUF protein family with distinct RNA binding specificity. RNA 2008, 14, 1550–1557. [Google Scholar] [CrossRef] [PubMed]
- Bachorik, J.L.; Kimble, J. Redundant control of the Caenorhabditis elegans sperm/oocyte switch by PUF-8 and FBF-1, two distinct PUF RNA-binding proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 10893–10897. [Google Scholar] [CrossRef] [PubMed]
- Ariz, M.; Mainpal, R.; Subramaniam, K. Caenorhabditis elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev. Biol. 2009, 326, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Nolde, M.J.; Saka, N.; Reinert, K.L.; Slack, F.J. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′ UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev. Biol. 2007, 305, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Forbes, A.; Lehmann, R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 1998, 125, 679–690. [Google Scholar] [PubMed]
- Asaoka-Taguchi, M.; Yamada, M.; Nakamura, A.; Hanyu, K.; Kobayashi, S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1999, 1, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Parisi, M.; Lin, H. The Drosophila pumilio gene encodes two functional protein isoforms that play multiple roles in germline development, gonadogenesis, oogenesis and embryogenesis. Genetics 1999, 153, 235–250. [Google Scholar] [PubMed]
- Mak, W.; Fang, C.; Holden, T.; Dratver, M.B.; Lin, H. An important role of Pumilio 1 in regulating the development of the mammalian female germline 1. Biol. Reprod. 2016, 94, 134. [Google Scholar] [CrossRef] [PubMed]
- Quenault, T.; Lithgow, T.; Traven, A. PUF proteins: Repression, activation and mRNA localization. Trends Cell Biol. 2011, 21, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.M.; Hong, E.L.; Amundsen, C.; Balakrishnan, R.; Binkley, G.; Chan, E.T.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; et al. Saccharomyces genome database: The genomics resource of budding yeast. Nucleic Acids Res. 2011, 40, D700–D705. [Google Scholar] [CrossRef] [PubMed]
- Goldstrohm, A.C.; Hook, B.A.; Seay, D.J.; Wickens, M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006, 13, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Olivas, W.; Parker, R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 2000, 19, 6602–6611. [Google Scholar] [CrossRef] [PubMed]
- Yosefzon, Y.; Koh, Y.Y.; Chritton, J.J.; Lande, A.; Leibovich, L.; Barziv, L.; Petzold, C.; Yakhini, Z.; Mandel-Gutfreund, Y.; Wickens, M.; et al. Divergent RNA binding specificity of yeast Puf2p. RNA 2011, 17, 1479–1488. [Google Scholar] [CrossRef] [PubMed]
- Machin, N.A.; Lee, J.M.; Barnes, G. Microtubule stability in budding yeast: Characterization and dosage suppression of a benomyl-dependent tubulin mutant. Mol. Biol. Cell 1995, 6, 1241–1259. [Google Scholar] [CrossRef] [PubMed]
- Jalal Kiani, S.; Taheri, T.; Rafati, S.; Samimi-Rad, K. PUF proteins: Cellular functions and potential applications. Curr. Protein Pept. Sci. 2017, 18, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.U.; Kim, M.J.; Paek, K.H. Arabidopsis Pumilio protein APUM5 suppresses cucumber mosaic virus infection via direct binding of viral RNAs. Proc. Natl. Acad. Sci. USA 2013, 110, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.U.; Paek, K.H. APUM5, encoding a Pumilio RNA binding protein, negatively regulates abiotic stress responsive gene expression. BMC Plant Biol. 2014, 14, 75. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Nakabayashi, K.; Ding, J.; He, F.; Bentsink, L.; Soppe, W.J. Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 2014, 26, 4362–4375. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Kerstetter, R.A.; Irish, V.F. APUM23, a PUF family protein, functions in leaf development and organ polarity in Arabidopsis. J. Exp. Bot. 2014, 65, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, T.; Abbasi, N.; Kim, H.S.; Kim, H.B.; Park, N.I.; Park, G.T.; Oh, S.A.; Park, S.K.; Muench, D.G.; Choi, Y.; et al. An arabidopsis divergent Pumilio protein, APUM24, Is essential for embryogenesis and required for faithful pre-rRNA processing. Plant J. 2017, 92, 1092–1105. [Google Scholar] [CrossRef] [PubMed]
- Weidmann, C.A.; Goldstrohm, A.C. Drosophila Pumilio protein contains multiple autonomous repression domains that regulate mRNAs independently of Nanos and brain tumor. Mol. Cell. Biol. 2012, 32, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Racher, H.; Hansen, D. PUF-8, a Pumilio homolog, inhibits the proliferative fate in the Caenorhabditis elegans germline. G3 Genes Genomes Genet. 2012, 2, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Hubstenberger, A.; Cameron, C.; Shtofman, R.; Gutman, S.; Evans, T.C. A network of PUF proteins and Ras signaling promote mRNA repression and oogenesis in Caenorhabditis elegans. Dev. Biol. 2012, 366, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Salvetti, A.; Rossi, L.; Lena, A.; Batistoni, R.; Deri, P.; Rainaldi, G.; Locci, M.T.; Evangelista, M.; Gremigni, V. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development 2005, 132, 1863–1874. [Google Scholar] [CrossRef] [PubMed]
- Goldstrohm, A.C.; Seay, D.J.; Hook, B.A.; Wickens, M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 2007, 282, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Herskowitz, I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev. 1988, 52, 536–553. [Google Scholar] [PubMed]
- Zhu, D.; Stumpf, C.R.; Krahn, J.M.; Wickens, M.; Hall, T.M.T. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc. Natl. Acad. Sci. USA 2009, 106, 20192–20197. [Google Scholar] [CrossRef] [PubMed]
- Wilinski, D.; Qiu, C.; Lapointe, C.P.; Nevil, M.; Campbell, Z.T.; Hall, T.M.T.; Wickens, M. RNA regulatory networks diversified through curvature of the PUF protein scaffold. Nat. Commun. 2015, 6, 8213. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; McCann, K.L.; Qiu, C.; Gonzalez, L.E.; Baserga, S.J.; Hall, T.M.T. Nop9 is a PUF-like protein that prevents premature cleavage to correctly process pre-18S rRNA. Nat. Commun. 2016, 7, 13085. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.S.; Krause, S.A.; McGhie, J.; Gray, J.V. Mpt5p, a stress tolerance-and lifespan-promoting PUF protein in Saccharomyces cerevisiae, acts upstream of the cell wall integrity pathway. Eukaryot. Cell 2007, 6, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Ye, W.; Situ, J.; Chen, Y.; Yang, X.; Kong, G.; Liu, Y.; Tinashe, R.J.; Xi, P.; Wang, Y.; et al. A PUF RNA-binding protein encoding gene PlM90 regulates the sexual and asexual life stages of the litchi downy blight pathogen Peronophythora litchii. Fungal Genet. Biol. 2017, 98, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Li, X.; Ning, G.; Miao, J.; Cui, L. The RNA-binding protein Puf1 functions in the maintenance of gametocytes in Plasmodium falciparum. J. Cell Sci. 2016, 129, 3144–3152. [Google Scholar] [PubMed]
- Narita, R.; Takahasi, K.; Murakami, E.; Hirano, E.; Yamamoto, S.P.; Yoneyama, M.; Kato, H.; Fujita, T. A novel function of human Pumilio proteins in cytoplasmic sensing of viral infection. PLoS Pathog. 2014, 10, e1004417. [Google Scholar] [CrossRef] [PubMed]
- Reichel, M.; Liao, Y.; Rettel, M.; Ragan, C.; Evers, M.; Alleaume, A.M.; Horos, R.; Hentze, M.W.; Preiss, T.; Millar, A.A. In planta determination of the mRNA-binding proteome of Arabidopsis etiolated seedlings. Plant Cell 2016, 28, 2435–3452. [Google Scholar] [CrossRef] [PubMed]
- Cooke, A.; Prigge, A.; Opperman, L.; Wickens, M. Targeted translational regulation using the PUF protein family scaffold. Proc. Natl. Acad. Sci. USA 2011, 108, 15870–15875. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, N.; Park, Y.I.; Choi, S.B. Pumilio Puf domain RNA-binding proteins in Arabidopsis. Plant Signal. Behav. 2011, 6, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Singer, R.H.; Gu, W. Translation of ASH1 mRNA is repressed by PUF6p–Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 2008, 22, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Luitjens, C.; Gallegos, M.; Kraemer, B.; Kimble, J.; Wickens, M. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 2000, 14, 2596–2609. [Google Scholar] [CrossRef] [PubMed]
- Kaye, J.A.; Rose, N.C.; Goldsworthy, B.; Goga, A.; Noelle, D.L. A 3′ UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron 2009, 61, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Hook, B.; Pan, G.; Kershner, A.M.; Merritt, C.; Seydoux, G.; Thomson, J.A.; Wickens, M.; Kimble, J. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007, 3, e233. [Google Scholar] [CrossRef] [PubMed]
- Moore, F.L.; Jaruzelska, J.; Fox, M.S.; Urano, J.; Firpo, M.T.; Turek, P.J.; Dorfman, D.M.; Pera, R.A. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc. Natl. Acad. Sci. USA 2003, 100, 538–543. [Google Scholar] [CrossRef] [PubMed]


| Organisms | PUF Family Member | Target mRNA | Binding Motif | Biological Function | References |
|---|---|---|---|---|---|
| Caenorhabditis elegans | FBF | Gld-1, Fem3 | 5′-UGUGCCAUA-3′, 5′-UGUGUCAUU-3′ | Maintenance of stem cell proliferation; the hermaphroditic switch between spermatogenesis and oogenesis; adaptation in the AWC chemosensory neuron. | [29,48,49] |
| PUF-5 | HIS3 (a reporter gene), obr-3, cpi-2, srm-6, fog-1, srz-10, C17H11 | 5′-CYCUGUAYYYUGU-3′ | Oocyte maturation; nuclear enlargement; yolk uptake; early embryogenesis | [49,50] | |
| PUF-6 | HIS3 (a reporter gene) | 5′-CYCUGUAYYYUGU-3′ | Primordial germ cell development | [49,50] | |
| PUF-7 | HIS3 (a reporter gene) | 5′-CYCUGUAYYYUGU-3′ | Primordial germ cell development | [49,50] | |
| PUF-8 | Unknown | Unknown | Hermaphrodite spermoocyte switch; Germ-Line Proliferation | [49,51,52] | |
| PUF-9 | Unknown | Unknown | Differentiation of epidermal stem cells at the larval-to-adult transition | [49,53] | |
| Cryptococcus neoformans | PUM1 | Znf2 | 5′-UGUACAUA-3′ | Hyphal morphogenesis of sexual development | [41] |
| Drosophila melanogaster | PUMILIO | hbNRE; hunchback; cyclin B; eIF4E; Bicoid; para | Nanos response element | Anterior patterning system; mitotic arrest of primordial germ cells; maintenance of germline stem cells; primordial follicle pool; gonadogenesis; oogenesis; neuronal function; sodium current in motoneurons | [15,21,40,54,55,56,57,58] |
| Saccharomyces cerevisiae | MPT5 | HO | Nanos response element | Mating-type switching; Lifespan | [23,59] |
| PUF4 | HO | Nanos response element | Lifespan | [1,59,60] | |
| PUF3 | COX17 | 5′-UGUAUAUAU-3′ | Mitochondrial biogenesis and motility; thermotolerance; hyperosmotic stress resistance | [32,59,61] | |
| PUF2 | Unknown | 5′-UAAUAAUUW-3′ | Binds mRNAs encoding membrane-associated proteins | [59,62] | |
| PUF1/JSN1 | Unknown | Unknown | A high copy suppressor of certain tubulin mutations | [59,63] | |
| PUF6 | ASH1 | 5′-UUGU-3′ motif | Mating-type switching; protein/peptide accumulation | [12,59] | |
| Xenopus | XPum2 | Xenopus cyclin B1 | 5′-UGUAAAUA-3′ | Oocyte maturation | [22,25,64] |
| Arabidopsis thaliana | APUM1-6 | FASCIATA-2, CLAVATA-1 and ZWILLE⁄PINHEAD | 5′UGUANAUA | shoot meristem organization, stem cell maintenance and maintenance of cellular organization of apical meristems | [42] |
| APUM5 | CMV tripartite RNA 3′UTR regions | 5′-UGUAAUA-3′; 5′-UGUAGUA-3′; 5′-UGUACAUAAUA-3′ | Defensive repressor of Cucumber mosaic virus (CMV) infection | [65] | |
| APUM5 | RAB18 COR15 RD22 DREB2A | 5′-UGUA-3′ | Abiotic stress response | [66] | |
| APUM9 and APUM11 | Unknown | Unknown | Seed dormancy | [67] | |
| APUM23 | Unknown | Unknown | Leaf development and organ polarity; Processing and/or degradation of 35S pre-rRNA and rRNA maturation by-products | [4,68] | |
| APUM24 | 7S pre-rRNA; ITS2 | Unknown | rRNA processing and early embryogenesis | [69] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Ogé, L.; Perez-Garcia, M.-D.; Hamama, L.; Sakr, S. The PUF Protein Family: Overview on PUF RNA Targets, Biological Functions, and Post Transcriptional Regulation. Int. J. Mol. Sci. 2018, 19, 410. https://doi.org/10.3390/ijms19020410
Wang M, Ogé L, Perez-Garcia M-D, Hamama L, Sakr S. The PUF Protein Family: Overview on PUF RNA Targets, Biological Functions, and Post Transcriptional Regulation. International Journal of Molecular Sciences. 2018; 19(2):410. https://doi.org/10.3390/ijms19020410
Chicago/Turabian StyleWang, Ming, Laurent Ogé, Maria-Dolores Perez-Garcia, Latifa Hamama, and Soulaiman Sakr. 2018. "The PUF Protein Family: Overview on PUF RNA Targets, Biological Functions, and Post Transcriptional Regulation" International Journal of Molecular Sciences 19, no. 2: 410. https://doi.org/10.3390/ijms19020410
APA StyleWang, M., Ogé, L., Perez-Garcia, M. -D., Hamama, L., & Sakr, S. (2018). The PUF Protein Family: Overview on PUF RNA Targets, Biological Functions, and Post Transcriptional Regulation. International Journal of Molecular Sciences, 19(2), 410. https://doi.org/10.3390/ijms19020410