Substrate Recognition and Specificity of Chitin Deacetylases and Related Family 4 Carbohydrate Esterases
Abstract
:1. Carbohydrate Esterases and the CE4 Family
2. Substrates of CE4 Family Enzymes
2.1. Chitin, Chitosan, and Their Oligomers
2.2. Peptidoglycan
2.3. Acetylxylan
2.4. β-1,6-Glucan
3. CE4 Enzymes Active on Chitooligosaccharides and Their Substrate Specificities
3.1. Chitin Deacetylases (CDAs)
3.1.1. Fungal Chitin Deacetylases
3.1.2. Bacterial Chitin Deacetylases
3.2. Peptidoglycan Deacetylases
3.2.1. GlcNAc Peptidoglycan Deacetylases
3.2.2. MurNAc Peptidoglycan Deacetylases
3.3. Putative Polysaccharide Deacetylases (PPda)
3.4. Acetylxylan Esterases
3.5. Poly-β-1,6-GlcNAc de-N-acetylase
4. Structural and Sequence Features of CE4 Enzymes Active on Chitooligosaccharides
4.1. Domain Organization
4.2. X-ray Structures
4.3. Multiple Sequence Alignment of the CE4 Domain
4.4. NodB Domain and Active Site Conserved Motifs
5. Substrate Recognition and Catalysis
5.1. Catalytic Mechanism
5.2. Substrate Recognition and Specificity
5.2.1. The Case of VcCDA: Enzyme·Substrate Complexes Show an Induced Fit Mechanism
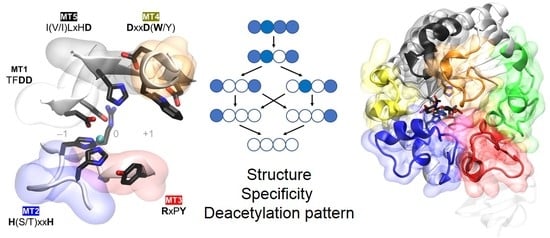
5.2.2. Determinants of Substrate Specificity: The Subsite Capping Model
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AXE | Acetylxylan estarase |
| An | (GlcNAc)n |
| CDA | Chitin deacetylase |
| COS | Chitooligosaccharides |
| Dn | (GlcNH2)n |
| DA | Degree of acetylation |
| DP | Degree of polymerization |
| GlcNAc | N-acetylglucosamine |
| GlcNH2 | Glucosamine |
| MurNAc | N-acetylmuramic acid |
| PA | Pattern of acetylation |
| PDB | Protein data bank |
| PG | Peptidoglycan |
| PNAG | Poly-β-1,6-N-acetyl-d-glucosamine |
| PPda | Putative polysaccharide deacetylase |
References
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.M.; Nascimento, A.S.; Polikarpov, I. Structural diversity of carbohydrate esterases. Biotechnol. Res. Innov. 2017, 1, 35–51. [Google Scholar] [CrossRef]
- Zhao, Y.; Park, R.D.; Muzzarelli, R.A.A. Chitin deacetylases: Properties and applications. Mar. Drugs 2010, 8, 24–46. [Google Scholar] [CrossRef] [PubMed]
- Biely, P. Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnol. Adv. 2012, 30, 1575–1588. [Google Scholar] [CrossRef] [PubMed]
- Sobhanifar, S.; King, D.T.; Strynadka, N.C.J. Fortifying the wall: Synthesis, regulation and degradation of bacterial peptidoglycan. Curr. Opin. Struct. Biol. 2013, 23, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Caufrier, F.; Martinou, A.; Dupont, C.; Bouriotis, V. Carbohydrate esterase family 4 enzymes: Substrate specificity. Carbohydr. Res. 2003, 338, 687–692. [Google Scholar] [CrossRef]
- John, M.; Rohrig, H.; Schmidt, J.; Wieneke, U.; Schell, J. Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc. Natl. Acad. Sci. USA 1993, 90, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Ghormade, V.; Kulkarni, S.; Doiphode, N.; Rajamohanan, P.R.; Deshpande, M.V. Chitin deacetylase: A comprehensive account on its role in nature and its biotechnological applications. In Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology; Formatex: Badajoz, Spain, 2010; pp. 1054–1066. [Google Scholar]
- Araki, Y.; Ito, E. A pathway of chitosan formation in mucor rouxii enzymatic deacetylation of chitin. Eur. J. Biochem. 1975, 55, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ju, W.; Jo, G.; Jung, W.; Park, R. Perspectives of chitin deacetylase research. Biotechnol. Biopolym. 2011, 131–145. [Google Scholar] [CrossRef]
- Hoßbach, J.; Bußwinkel, F.; Kranz, A.; Wattjes, J.; Cord-Landwehr, S.; Moerschbacher, B.M. A chitin deacetylase of Podospora anserina has two functional chitin binding domains and a unique mode of action. Carbohydr. Polym. 2018, 183, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gay, L.M.; Tuveng, T.R.; Agger, J.W.; Westereng, B.; Mathiesen, G.; Horn, S.J.; Vaaje-Kolstad, G.; van Aalten, D.M.F.; Eijsink, V.G.H. Structure and function of a broad-specificity chitin deacetylase from Aspergillus nidulans FGSC A4. Sci. Rep. 2017, 7, 1746. [Google Scholar] [CrossRef] [PubMed]
- Tsigos, I.; Martinou, A.; Kafetzopoulos, D.; Bouriotis, V. Chitin deacetylases: New, versatile tools in biotechnology. Trends Biotechnol. 2000, 18, 305–312. [Google Scholar] [CrossRef]
- Peniche Covas, C.A.; Argüelles-Monal, W.; Goycoolea, F.M. Chitin and chitosan: Major sources, properties and applications. In Monomers, Polymers and Composites from Renewable Resources; Elsevier Science: Amsterdam, The Netherlands, 2008; Volume 1, pp. 517–542. [Google Scholar]
- Karrer, P.; Hofmann, A. Über den enzymatischen Abbau von Chitin und Chitosan I. Helv. Chim. Acta 1929, 12, 616–637. [Google Scholar] [CrossRef]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Dutta, P.K.; Duta, J.; Tripathi, V.S. Chitin and Chitosan: Chemistry, properties and applications. J. Sci. Ind. Res. 2004, 63, 20–31. [Google Scholar]
- Kumirska, J.; Czerwicka, M.; Kaczyński, Z.; Bychowska, A.; Brzozowski, K.; Thöming, J.; Stepnowski, P. Application of spectroscopic methods for structural analysis of chitin and chitosan. Mar. Drugs 2010, 8, 1567–1636. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, G.S.; Kaur, S.; Brar, S.K.; Verma, M. Green synthesis approach: Extraction of chitosan from fungus mycelia. Crit. Rev. Biotechnol. 2013, 33, 379–403. [Google Scholar] [CrossRef] [PubMed]
- Hoell, I.A.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Structure and function of enzymes acting on chitin and chitosan. Biotechnol. Genet. Eng. Rev. 2010, 27, 331–366. [Google Scholar] [CrossRef]
- Xia, W.; Liu, P.; Zhang, J.; Chen, J. Biological activities of chitosan and chitooligosaccharides. Food Hydrocoll. 2011, 25, 170–179. [Google Scholar] [CrossRef]
- Yu, R.; Liu, W.; Li, D.; Zhao, X.; Ding, G.; Zhang, M.; Ma, E.; Zhu, K.Y.; Li, S.; Moussian, B.; et al. Helicoidal organization of chitin in the cuticle of the migratory locust requires the function of the chitin deacetylase2 enzyme (LmCDA2). J. Biol. Chem. 2016, 291, 24352–24363. [Google Scholar] [CrossRef] [PubMed]
- Winkler, A.J.; Dominguez-Nuñez, J.A.; Aranaz, I.; Poza-Carrión, C.; Ramonell, K.; Somerville, S.; Berrocal-Lobo, M. Short-chain chitin oligomers: Promoters of plant growth. Mar. Drugs 2017, 15, 40. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Min, M.; Du, N.; Gu, Y.; Hode, T.; Naylor, M.; Chen, D.; Nordquist, R.E.; Chen, W.R. Chitin, chitosan, and glycated chitosan regulate immune responses: The novel adjuvants for cancer vaccine. Clin. Dev. Immunol. 2013, 2013, 387023. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Sharon, N. Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R., Esko, J., Freeze, H., Stanley, P., Bertozzi, C.R., Hart, G., Etzler, M.E., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009. [Google Scholar]
- Park, B.K.; Kim, M.M. Applications of chitin and its derivatives in biological medicine. Int. J. Mol. Sci. 2010, 11, 5152–5164. [Google Scholar] [CrossRef] [PubMed]
- Muanprasat, C.; Chatsudthipong, V. Chitosan oligosaccharide: Biological activities and potential therapeutic applications. Pharmacol. Ther. 2017, 170, 80–97. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Blanot, D.; De Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [PubMed]
- Mengin-Lecreulx, D.; Lemaitre, B. Structure and metabolism of peptidoglycan and molecular requirements allowing its detection by the Drosophila innate immune system. J. Endotoxin Res. 2005, 11, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Johannsen, L. Biological properties of bacterial peptidoglycan. Apmis 1993, 101, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, R.; Chevalier, G.; Eberl, G.; Gomperts Boneca, I. The biology of bacterial peptidoglycans and their impact on host immunity and physiology. Cell. Microbiol. 2014, 16, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Van Heijenoort, J. Peptidoglycan hydrolases of escherichia coli. Microbiol. Mol. Biol. Rev. 2011, 75, 636–663. [Google Scholar] [CrossRef] [PubMed]
- Clarke, A.J. Chemical biology of peptidoglycan acetylation and deacetylation. Bioorg. Chem. 2014, 54, 44–50. [Google Scholar]
- Boneca, I.G. The role of peptidoglycan in pathogenesis. Curr. Opin. Microbiol. 2005, 8, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Tomasz, A. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 2000, 275, 20496–20501. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Tomasz, A. Peptidoglycan N-Acetylglucosamine Deacetylase, a Putative Virulence Factor in Streptococcus pneumoniae. Infect. Immun. 2002, 70, 7176–7178. [Google Scholar] [CrossRef] [PubMed]
- Popper, Z.A.; Michel, G.; Hervé, C.; Domozych, D.S.; Willats, W.G.T.; Tuohy, M.G.; Kloareg, B.; Stengel, D.B. Evolution and Diversity of Plant Cell Walls: From Algae to Flowering Plants. Annu. Rev. Plant Biol. 2011, 62, 567–590. [Google Scholar] [CrossRef] [PubMed]
- Ebringerová, A.; Hromádková, Z.; Heinze, T. Hemicellulose. In Polysaccharides I: Structure, Characterization and Use; Heinze, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–67. ISBN 978-3-540-31583-4. [Google Scholar]
- Puchart, V.; Biely, P. Redistribution of acetyl groups on the non-reducing end xylopyranosyl residues and their removal by xylan deacetylases. Appl. Microbiol. Biotechnol. 2015, 99, 3865–3873. [Google Scholar] [CrossRef] [PubMed]
- Biely, P.; Cziszárová, M.; Uhliariková, I.; Agger, J.W.; Li, X.L.; Eijsink, V.G.H.; Westereng, B. Mode of action of acetylxylan esterases on acetyl glucuronoxylan and acetylated oligosaccharides generated by a GH10 endoxylanase. Biochim. Biophys. Acta 2013, 1830, 5075–5086. [Google Scholar] [CrossRef] [PubMed]
- Adesioye, F.A.; Makhalanyane, T.P.; Biely, P.; Cowan, D.A. Phylogeny, classification and metagenomic bioprospecting of microbial acetyl xylan esterases. Enzym. Microb. Technol. 2016, 93–94, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; Vik, Å.; Friedman, L.; Kolter, R. Biofilms: The matrix revisited. Trends Microbiol. 2005, 13, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Vu, B.; Chen, M.; Crawford, R.J.; Ivanova, E.P. Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 2009, 14, 2535–2554. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Rice, J.D.; Goller, C.; Pannuri, A.; Taylor, J.; Meisner, J.; Beveridge, T.J.; Preston, J.F.; Romeo, T. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-d-glucosamine. J. Bacteriol. 2008, 190, 3670–3680. [Google Scholar] [CrossRef] [PubMed]
- Little, D.J.; Milek, S.; Bamford, N.C.; Ganguly, T.; Difrancesco, B.R.; Nitz, M.; Deora, R.; Howell, P.L. The protein BpsB is a poly-β-1,6-N-acetyl-d-glucosamine deacetylase required for biofilm formation in Bordetella bronchiseptica. J. Biol. Chem. 2015, 290, 22827–22840. [Google Scholar] [CrossRef] [PubMed]
- Chibba, A.; Poloczek, J.; Little, D.J.; Howell, P.L.; Nitz, M. Synthesis and evaluation of inhibitors of E. coli PgaB, a polysaccharide de-N-acetylase involved in biofilm formation. Org. Biomol. Chem. 2012, 10, 7103–7107. [Google Scholar] [CrossRef] [PubMed]
- Martinou, A.; Bouriotis, V.; Stokke, B.T.; Vårum, K.M. Mode of action of chitin deacetylase from Mucor rouxii on partially N-acetylated chitosans. Carbohydr. Res. 1998, 311, 71–78. [Google Scholar] [CrossRef]
- Tsigos, I.; Zydowicz, N.; Martinou, A.; Domard, A.; Bouriotis, V. Mode of action of chitin deacetylase from Mucor rouxii on N-acetylchitooligosaccharides. Eur. J. Biochem. 1999, 261, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Tokuyasu, K.; Mitsutomi, M.; Yamaguchi, I.; Hayashi, K.; Mori, Y. Recognition of chitooligosaccharides and their N-acetyl groups by putative subsites of chitin deacetylase from a Deuteromycete, Colletotrichum lindemuthianum. Biochemistry 2000, 39, 8837–8843. [Google Scholar] [CrossRef] [PubMed]
- Hekmat, O.; Tokuyasu, K.; Withers, S.G. Subsite structure of the endo-type chitin deacetylase from a deuteromycete, Colletotrichum lindemuthianum: An investigation using steady-state kinetic analysis and MS. Biochem. J. 2003, 374, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.G.; Specht, C.A.; Donlin, M.J.; Lodge, J.K. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell 2007, 6, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; Tanaka, K.; Matsuda, H.; Kawamukai, M. Cda1+, encoding chitin deacetylase is required for proper spore formation in Schizosaccharomyces pombe. FEBS Lett. 2005, 579, 2737–2743. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, I.A.; Gurr, S.J. Chitosan Mediates Germling Adhesion in Magnaporthe oryzae and Is Required for Surface Sensing and Germling Morphogenesis. PLoS Pathog. 2016, 12, 1–34. [Google Scholar] [CrossRef] [PubMed]
- White, S.; McIntyre, M.; Berry, D.R.; McNeil, B. The autolysis of industrial filamentous fungi. Crit. Rev. Biotechnol. 2002, 22, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vallet, A.; Mesters, J.R.; Thomma, B.P.H.J. The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 2015, 39, 171–183. [Google Scholar] [CrossRef] [PubMed]
- El Gueddari, N.E.; Rauchhaus, U.; Moerschbacher, B.M.; Deising, H.B. Developmentally regulated conversion of surface-exposed chitin to chi- tosan in cell walls of plant pathogenic fungi. New Phytol. 2002, 156, 103–112. [Google Scholar] [CrossRef]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J.; et al. Chitin-induced dimerization activates a plant immune receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Cord-Landwehr, S.; Melcher, R.L.J.; Kolkenbrock, S.; Moerschbacher, B.M. A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci. Rep. 2016, 6, 38018. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.L.; Bartnicki-Garcia, S. Chitosan synthesis by the tandem action of chitin synthetase and chitin deacetylase from mucor rouxii. Biochemistry 1984, 23, 1065–1073. [Google Scholar] [CrossRef]
- Kafetzopoulos, D.; Martinou, A.; Bouriotis, V. Bioconversion of chitin to chitosan: Purification and characterization of chitin deacetylase from Mucor rouxii. Proc. Natl. Acad. Sci. USA 1993, 90, 2564–2568. [Google Scholar] [CrossRef] [PubMed]
- Blair, D.E.; Hekmat, O.; Schüttelkopf, A.W.; Shrestha, B.; Tokuyasu, K.; Withers, S.G.; van Aalten, D.M. Structure and mechanism of chitin deacetylase from the fungal pathogen colletotrichium lindemuthianum. Biochemistry 2006, 45, 9416–9426. [Google Scholar] [CrossRef] [PubMed]
- Andrés, E.; Albesa-Jové, D.; Biarnés, X.; Moerschbacher, B.M.; Guerin, M.E.; Planas, A. Structural basis of chitin oligosaccharide deacetylation. Angew. Chem. 2014, 53, 6882–6887. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Sugiyama, K.; Sakaki, Y.; Hakamata, W.; Park, S.Y.; Nishio, T. Structure-based analysis of domain function of chitin oligosaccharide deacetylase from Vibrio parahaemolyticus. FEBS Lett. 2015, 589, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Tuveng, T.R.; Rothweiler, U.; Udatha, G.; Vaaje-Kolstad, G.; Smalås, A.; Eijsink, V.G.H. Structure and function of a CE4 deacetylase isolated from a marine environment. PLoS ONE 2017, 12, e0187544. [Google Scholar] [CrossRef] [PubMed]
- Blair, D.E.; Schuttelkopf, A.W.; MacRae, J.I.; van Aalten, D.M.F. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl. Acad. Sci. USA 2005, 102, 15429–15434. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.D.; Urch, J.E.; Ten Cate, J.M.; Rao, V.A.; Van Aalten, D.M.F.; Crielaard, W. Streptococcus mutans SMU.623c codes for a functional, metal-dependent polysaccharide deacetylase that modulates interactions with salivary agglutinin. J. Bacteriol. 2009, 91, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Fadouloglou, V.E.; Balomenou, S.; Aivaliotis, M.; Kotsifaki, D.; Arnaouteli, S.; Tomatsidou, A.; Efstathiou, G.; Kountourakis, N.; Miliara, S.; Griniezaki, M.; et al. Unusual α-Carbon Hydroxylation of Proline Promotes Active-Site Maturation. J. Am. Chem. Soc. 2017, 139, 5330–5337. [Google Scholar] [CrossRef] [PubMed]
- Blair, D.E.; Van Aalten, D.M.F. Structures of Bacillus subtilis PdaA, a family 4 carbohydrate esterase, and a complex with N-acetyl-glucosamine. FEBS Lett. 2004, 570, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Oberbarnscheidt, L.; Taylor, E.J.; Davies, G.J.; Gloster, T.M. Structure of a Carbohydrate Esterase from Bacillus anthracis. Proteins Struct. Funct. Bioinform. 2007, 66, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Fadouloglou, V.E.; Kapanidou, M.; Agiomirgianaki, A.; Arnaouteli, S.; Bouriotis, V.; Glykos, N.M.; Kokkinidis, M. Structure determination through homology modelling and torsion-angle simulated annealing: Application to a polysaccharide deacetylase from Bacillus cereus. Acta Crystallogr. Sect. D 2013, 69, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Arnaouteli, S.; Giastas, P.; Andreou, A.; Tzanodaskalaki, M.; Aldridge, C.; Tzartos, S.J.; Vollmer, W.; Eliopoulos, E.; Bouriotis, V. Two putative polysaccharide deacetylases are required for osmotic stability and cell shape maintenance in Bacillus anthracis. J. Biol. Chem. 2015, 290, 13465–13478. [Google Scholar] [CrossRef] [PubMed]
- Strunk, R.J.; Piemonte, K.M.; Petersen, N.M.; Koutsioulis, D.; Bouriotis, V.; Perry, K.; Cole, K.E. Structure determination of BA0150, a putative polysaccharide deacetylase from Bacillus anthracis. Acta Crystallogr. Sect. F 2014, 70, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Urch, J.E.; Hurtado-Guerrero, R.; Brosson, D.; Liu, Z.; Eijsink, V.G.H.; Texier, C.; Van Aalten, D.M.F. Structural and functional characterization of a putative polysaccharide deacetylase of the human parasite Encephalitozoon cuniculi. Protein Sci. 2009, 18, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.J.; Gloster, T.M.; Turkenburg, J.P.; Vincent, F.; Brzozowski, A.M.; Dupont, C.; Shareck, F.; Centeno, M.S.J.; Prates, J.A.M.; Puchart, V.; et al. Structure and activity of two metal ion-dependent acetylxylan esterases involved in plant cell wall degradation reveals a close similarity to peptidoglycan deacetylases. J. Biol. Chem. 2006, 281, 10968–10975. [Google Scholar] [CrossRef] [PubMed]
- Little, D.J.; Poloczek, J.; Whitney, J.C.; Robinson, H.; Nitz, M.; Howell, P.L. The structure- and metal-dependent activity of Escherichia coli PgaB provides insight into the partial de-N-acetylation of poly-β-1,6-N-acetyl-d-glucosamine. J. Biol. Chem. 2012, 287, 31126–31137. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Noguchi, H.; Yoshida, H.; Park, S.Y.; Tame, J.R.H. The structure of the deacetylase domain of Escherichia coli PgaB, an enzyme required for biofilm formation: A circularly permuted member of the carbohydrate esterase 4 family. Acta Crystallogr. Sect. D 2013, 69, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Little, D.J.; Bamford, N.C.; Pokrovskaya, V.; Robinson, H.; Nitz, M.; Howell, P.L. Structural basis for the De-N-acetylation of poly-β-1,6-N-acetyl-d-glucosamine in gram-positive bacteria. J. Biol. Chem. 2014, 289, 35907–35917. [Google Scholar] [CrossRef] [PubMed]
- Kauss, H.; Bauch, B. Chitin deacetylase from Colletotrichum lindemuthianum. Methods Enzymol. 1988, 161, 518–523. [Google Scholar]
- Tokuyasu, K.; Ono, H.; Ohnishi-Kameyama, M.; Hayashi, K.; Mori, Y. Deacetylation of chitin oligosaccharides of dp 2-4 by chitin deacetylase from Colletotrichum lindemuthianum. Carbohydr. Res. 1997, 303, 353–358. [Google Scholar] [CrossRef]
- Tokuyasu, K.; Ohnishi-Kameyama, M.; Hayashi, K. Purification and characterization of extracellular chitin deacetylase from Colletotrichum lindemuthianum. Biosci. Biotechnol. Biochem. 1996, 60, 1598–1603. [Google Scholar] [CrossRef] [PubMed]
- Tokuyasu, K.; Ono, H.; Hayashi, K.; Mori, Y. Reverse hydrolysis reaction of chitin deacetylase and enzymatic synthesis of β-d-GlcNAc-(1→4)-GlcN from chitobiose. Carbohydr. Res. 1999, 322, 26–31. [Google Scholar] [CrossRef]
- Tokuyasu, K.; Ono, H.; Mitsutomi, M.; Hayashi, K.; Mori, Y. Synthesis of a chitosan tetramer derivative, β-d-GlcNAc-(1→4)-β-d-GlcNAc-(1→4)-β-d-GlcNAc-(1→4)-d-GlcN through a partial N-acetylation reaction by chitin deacetylase. Carbohydr. Res. 2000, 325, 211–215. [Google Scholar] [CrossRef]
- Kang, L.X.; Liang, Y.X.; Ma, L.X. Novel characteristics of chitin deacetylase from Colletotrichum lindemuthianum: Production of fully acetylated chitooligomers, and hydrolysis of deacetylated chitooligomers. Process Biochem. 2014, 49, 1936–1940. [Google Scholar] [CrossRef]
- Reyes, F.; Calatayud, J.; Martinez, M.J. Endochitinase from Aspergillus nidulans implicated in the autolysis of its cell wall. FEMS Microbiol. Lett. 1989, 51, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Reyes, F.; Calatayud, J.; Vazquez, C.; Martínez, M.J. β-N-Acetylglucosaminidase from Aspergillus nidulans which degrades chitin oligomers during autolysis. FEMS Microbiol. Lett. 1990, 65, 83–87. [Google Scholar] [CrossRef]
- Emri, T.; Molnár, Z.; Szilágyi, M.; Pócsi, I. Regulation of autolysis in aspergillus nidulans. Appl. Biochem. Biotechnol. 2008, 151, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Alfonso, C.; Nuero, O.M.; Santamaría, F.; Reyes, F. Purification of a heat-stable chitin deacetylase from Aspergillus nidulans and its role in cell wall degradation. Curr. Microbiol. 1995, 30, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, M.; Upadhyaya, N.M.; Sperschneider, J.; Park, R.F.; Szabo, L.J.; Steffenson, B.; Ellis, J.G.; Dodds, P.N. Changing the game: Using integrative genomics to probe virulence mechanisms of the stem rust pathogen puccinia graminis f. sp. tritici. Front. Plant Sci. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.; Cord-Landwehr, S.; Singh, R.; Bernard, F.; Kolkenbrock, S.; El Gueddari, N.E.; Moerschbacher, B.M. A recombinant fungal chitin deacetylase produces fully defined chitosan oligomers with novel patterns of acetylation. Appl. Environ. Microbiol. 2016, 82, 6645–6655. [Google Scholar] [CrossRef] [PubMed]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Groenewald, J.Z.; Xu, J.; Crous, P.W. Pestalotiopsis revisited. Stud. Mycol. 2014, 79, 121–186. [Google Scholar] [CrossRef] [PubMed]
- Espagne, E.; Lespinet, O.; Malagnac, F.; Da Silva, C.; Jaillon, O.; Porcel, B.M.; Couloux, A.; Aury, J.-M.; Ségurens, B.; Poulain, J.; et al. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 2008, 9, R77. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Llorca, L.V.; Olivares-Bernabeu, C.; Salinas, J.; Jansson, H.-B.; Kolattukudy, P.E. Pre-penetration events in fungal parasitism of nematode eggs. Mycol. Res. 2002, 106, 499–506. [Google Scholar] [CrossRef]
- Manzanilla-Lopez, R.H.; Esteves, I.; Finetti-Sialer, M.M.; Hirsch, P.R.; Ward, E.; Devonshire, J.; Hidalgo-Diaz, L. Pochonia chlamydosporia: Advances and challenges to improve its performance as a biological control agent of sedentary endo-parasitic nematodes. J. Nematol. 2013, 45, 1–7. [Google Scholar] [PubMed]
- Larriba, E.; Jaime, M.D.L.A.; Carbonell-Caballero, J.; Conesa, A.; Dopazo, J.; Nislow, C.; Martín-Nieto, J.; Lopez-Llorca, L.V. Sequencing and functional analysis of the genome of a nematode egg-parasitic fungus, Pochonia chlamydosporia. Fungal Genet. Biol. 2014, 65, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Aranda-Martinez, A.; Grifoll-Romero, L.; Aragunde Pazos, H.; Enea, S.-V.; Biarnés, X.; Lopez-Llorca, L.V.; Planas, A. Expression and specificity of a chitin deacetylase catalytic domain from the nematophagous fungus Pochonia chlamydosporia potentially involved in pathogenicity. Sci. Rep. 2018, in press. [Google Scholar]
- Egelhoff, T.T.; Long, S.R. Rhizobium meliloti nodulation genes: Identification of nodDABC gene products, purification of nodA protein, and expression of nodA in Rhizobium meliloti. J. Bacteriol. 1985, 164, 591–599. [Google Scholar] [PubMed]
- Spaink, H.P.; Wijfjes, A.H.M.; der van Drift, K.M.G.M.; Haverkamp, J.; Thomas-Oates, J.E.; Lugtenberg, B.J.J. Structural identification of metabolites produced by the NodB and NodC proteins of Rhizobium leguminosarum. Mol. Microbiol. 1994, 13, 821–831. [Google Scholar] [CrossRef] [PubMed]
- Roche, P.; Maillet, F.; Plazanet, C.; Debellé, F.; Ferro, M.; Truchet, G.; Promé, J.C.; Dénarié, J. The common nodABC genes of Rhizobium meliloti are host-range determinants. Proc. Natl. Acad. Sci. USA 1996, 93, 15305–15310. [Google Scholar] [CrossRef] [PubMed]
- Chambon, R.; Pradeau, S.; Fort, S.; Cottaz, S.; Armand, S. High yield production of Rhizobium NodB chitin deacetylase and its use for in vitro synthesis of lipo-chitinoligosaccharide precursors. Carbohydr. Res. 2017, 442, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Hamer, S.N.; Cord-Landwehr, S.; Biarnés, X.; Planas, A.; Waegeman, H.; Moerschbacher, B.M.; Kolkenbrock, S. Enzymatic production of defined chitosan oligomers with a specific pattern of acetylation using a combination of chitin oligosaccharide deacetylases. Sci. Rep. 2015, 5, 8716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samain, E.; Chazalet, V.; Geremia, R.A. Production of O-acetylated and sulfated chitooligosaccharides by recombinant Escherichia coli strains harboring different combinations of nod genes. J. Biotechnol. 1999, 72, 33–47. [Google Scholar] [CrossRef]
- Keyhani, N.O.; Roseman, S. Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta 1999, 1473, 108–122. [Google Scholar] [CrossRef]
- Zobell, C.; Rittenberg, S. The occurrence and characteristics of chitinoclastic bacteria in the sea. J. Bacteriol. 1937, 35, 275–287. [Google Scholar]
- Meibom, K.L.; Li, X.B.; Nielsen, A.T.; Wu, C.-Y.; Roseman, S.; Schoolnik, G.K. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. USA 2004, 101, 2524–2529. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Roseman, S. The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. Proc. Natl. Acad. Sci. USA 2004, 101, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Ohishi, K.; Yamagishi, M.; Ohta, T.; Motosugi, M.; Izumida, H.; Sano, H.; Adachi, K.; Miwa, T. Purification and Properties of Two Deacetylases Produced by Vibrio alginolyticus H-8. Biosci. Biotechnol. Biochem. 1997, 61, 1113–1117. [Google Scholar] [CrossRef]
- Ohishi, K.; Murase, K.; Ohta, T.; Etoh, H. Cloning and sequencing of the deacetylase gene from Vibrio alginolyticus H-8. J. Biosci. Bioeng. 2000, 90, 561–563. [Google Scholar] [CrossRef]
- Kadokura, K.; Rokutani, A.; Yamamoto, M.; Ikegami, T.; Sugita, H.; Itoi, S.; Hakamata, W.; Oku, T.; Nishio, T. Purification and characterization of Vibrio parahaemolyticus extracellular chitinase and chitin oligosaccharide deacetylase involved in the production of heterodisaccharide from chitin. Appl. Microbiol. Biotechnol. 2007, 75, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Kadokura, K.; Sakamoto, Y.; Saito, K.; Ikegami, T.; Hirano, T.; Hakamata, W.; Oku, T.; Nishio, T. Production of a recombinant chitin oligosaccharide deacetylase from Vibrio parahaemolyticus in the culture medium of Escherichia coli cells. Biotechnol. Lett. 2007, 29, 1209–1215. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, L.X.; Wang, X.; Roseman, S. The chitin catabolic cascade in the marine bacterium Vibrio cholerae: Characterization of a unique chitin oligosaccharide deacetylase. Glycobiology 2007, 17, 1377–1387. [Google Scholar] [CrossRef] [PubMed]
- Jacquiod, S.; Franqueville, L.; Cécillon, S.; Vogel, T.M.; Simonet, P. Soil bacterial community shifts after Chitin enrichment: An integrative metagenomic approach. PLoS ONE 2013, 8, e79699. [Google Scholar] [CrossRef] [PubMed]
- Dsouza, M.; Taylor, M.W.; Turner, S.J.; Aislabie, J. Genomic and phenotypic insights into the ecology of Arthrobacter from Antarctic soils. BMC Genom. 2015, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Lonhienne, T.; Mavromatis, K.; Vorgias, C.E.; Buchon, L.; Gerday, C.; Bouriotis, V. Cloning, sequences, and characterization of two chitinase genes from the Antarctic Arthrobacter sp. strain TAD20: Isolation and partial characterization of the enzymes. J. Bacteriol. 2001, 183, 1773–1779. [Google Scholar] [CrossRef] [PubMed]
- Weiser, J.N. The pneumococcus: Why a commensal misbehaves. J. Mol. Med. 2010, 88, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Bui, N.K.; Turk, S.; Buckenmaier, S.; Stevenson-Jones, F.; Zeuch, B.; Gobec, S.; Vollmer, W. Development of screening assays and discovery of initial inhibitors of pneumococcal peptidoglycan deacetylase PgdA. Biochem. Pharmacol. 2011, 82, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Duport, C.; Jobin, M.; Schmitt, P. Adaptation in Bacillus cereus: From stress to disease. Front. Microbiol. 2016, 7, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Oba, S.; Araki, S.; Ito, E. Enzymatic deacetylation of N-acetylglucosamine residues in cell wall peptidoglycan. J. Biochem. 1980, 88, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Psylinakis, E.; Boneca, I.G.; Mavromatis, K.; Deli, A.; Hayhurst, E.; Foster, S.J.; Vårum, K.M.; Bouriotis, V. Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus anthracis. J. Biol. Chem. 2005, 280, 30856–30863. [Google Scholar] [CrossRef] [PubMed]
- Balomenou, S.; Fouet, A.; Tzanodaskalaki, M.; Couture-Tosi, E.; Bouriotis, V.; Boneca, I.G. Distinct functions of polysaccharide deacetylases in cell shape, neutral polysaccharide synthesis and virulence of Bacillus anthracis. Mol. Microbiol. 2013, 87, 867–883. [Google Scholar] [CrossRef] [PubMed]
- Mahowald, M.A.; Rey, F.E.; Seedorf, H.; Turnbaugh, P.J.; Fulton, R.S.; Wollam, A.; Shah, N.; Wang, C.; Magrini, V.; Wilson, R.K.; et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. USA 2009, 106, 5859–5864. [Google Scholar] [CrossRef] [PubMed]
- McKenney, P.T.; Driks, A.; Eichenberger, P. The Bacillus subtilis endospore: Assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 2012, 11, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, T.; Yamamoto, H.; Atrih, A.; Foster, S.J.; Sekiguchi, J.; Court, F.; Bank, W.; Sheffield, S.; Kingdom, U. A Polysaccharide Deacetylase Gene (pdaA) Is Required for Germination and for Production of Muramic δ-Lactam Residues in the Spore Cortex of Bacillus subtilis. J. Bacteriol. 2002, 184, 6007–6015. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, T.; Kitajima, T.; Sekiguchi, J. A polysaccharide deacetylase homologue, PdaA, in Bacillus subtilis acts as an N-acetylmuramic acid deacetylase in vitro. J. Bacteriol. 2005, 187, 1287–1292. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.E.; Bandyopadhyay, D.; Dean, A.M.; Linnstaedt, S.D.; Popham, D.L. Production of Muramic δ-Lactam in Bacillus subtilis Spore Peptidoglycan. J. Bacteriol. 2004, 186, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Żakowska, D.; Bartoszcze, M.; Niemcewicz, M.; Bielawska-Drózd, A.; Knap, J.; Cieślik, P.; Chomiczewski, K.; Kocik, J. Bacillus anthracis infections—New possibilities of treatment. Ann. Agric. Environ. Med. 2015, 22, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Fast, N.M. Microsporidia: Biology and evolution of highly reduced intracellular parasites. Annu. Rev. Microbiol. 2002, 56, 93–116. [Google Scholar] [CrossRef] [PubMed]
- Didier, E.S. Microsporidiosis: An emerging and opportunistic infection in humans and animals. Acta Trop. 2005, 94, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.; Morosoli, R.; Shareck, F.; Kluepfel, D. Production and secretion of proteins by streptomycetes. Crit. Rev. Biotechnol. 1995, 15, 13–39. [Google Scholar] [CrossRef] [PubMed]
- Biely, P. Microbial xylanolytic systems. Trends Biotechnol. 1985, 3, 286–290. [Google Scholar] [CrossRef]
- Vats-Mehta, S.; Bouvrette, P.; Shareck, F.; Morosoli, R.; Kluepfel, D. Cloning of a second xylanase-enconding gene of Streptomyces lividans 66. Gene 1996, 86, 119–122. [Google Scholar] [CrossRef]
- Shareck, F.; Biely, P.; Morosoli, R.; Kluepfel, D. Analysis of DNA flanking the xlnB locus of Streptomyces lividans reveals genes encoding acetyl xylan esterase and the RNA component of ribonuclease P. Gene 1995, 153, 105–109. [Google Scholar] [CrossRef]
- Biely, P.; Côté, G.L.; Kremnický, L.; Greene, R.V.; Dupont, C.; Kluepfel, D. Substrate specificity and mode of action of acetylxylan esterase from Streptomyces lividans. FEBS Lett. 1996, 396, 257–260. [Google Scholar] [CrossRef]
- Biely, P.; Mastihubová, M.; Côté, G.L.; Greene, R.V. Mode of action of acetylxylan esterase from Streptomyces lividans: A study with deoxy and deoxy-fluoro analogues of acetylated methyl β-d-xylopyranoside. Biochim. Biophys. Acta 2003, 1622, 82–88. [Google Scholar] [CrossRef]
- Uhliariková, I.; Vršanská, M.; McCleary, B.V.; Biely, P. Positional specifity of acetylxylan esterases on natural polysaccharide: An NMR study. Biochim. Biophys. Acta 2013, 1830, 3365–3372. [Google Scholar] [CrossRef] [PubMed]
- Morley, K.L.; Chauve, G.; Kazlauskas, R.; Dupont, C.; Shareck, F.; Marchessault, R.H. Acetyl xylan esterase-catalyzed deacetylation of chitin and chitosan. Carbohydr. Polym. 2006, 63, 310–315. [Google Scholar] [CrossRef]
- Tang, M.-C.; Nisole, A.; Dupont, C.; Pelletier, J.N.; Waldron, K.C. Chemical profiling of the deacetylase activity of acetyl xylan esterase A (AxeA) variants on chitooligosaccharides using hydrophilic interaction chromatography-mass spectrometry. J. Biotechnol. 2011, 155, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Puchart, V.; Gariépy, M.C.; Shareck, F.; Dupont, C. Identification of catalytically important amino acid residues of Streptomyces lividans acetylxylan esterase A from carbohydrate esterase family 4. Biochim. Biophys. Acta 2006, 1764, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Lamed, R.; Bayer, E.A. Cellulosomes from Clostridium thermocellum. Methods Enzymol. 1988, 160, 472–482. [Google Scholar]
- Smith, S.P.; Bayer, E.A. Insights into cellulosome assembly and dynamics: From dissection to reconstruction of the supramolecular enzyme complex. Curr. Opin. Struct. Biol. 2013, 23, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.C.; Fontes, C.M.; Gilbert, H.J.; Hazlewood, G.P.; Fernandes, T.H.; Ferreira, L.M. Homologous xylanases from Clostridium thermocellum: Evidence for bi-functional activity, synergism between xylanase catalytic modules and the presence of xylan-binding domains in enzyme complexes. Biochem. J. 1999, 342, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Biely, P.; Mastihubová, M.; Puchart, V. The vicinal hydroxyl group is prerequisite for metal activation of Clostridium thermocellum acetylxylan esterase. Biochim. Biophys. Acta 2007, 1770, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.; Moser, C.; Jensen, P.Ø.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef] [PubMed]
- Furkanur Rahaman Mizan, M.; Kabir Jahid, I.; Ha, S.-D. Microbial biofilms in seafood: A food-hygiene challenge. Food Microbiol. 2015, 49, 41–55. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, Z.; Palazzi, F.; Giardino, L.; Shalavi, S. Microbial biofilms in endodontic infections: An update review. Biomed. J. 2013, 36, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Gbejuade, H.O.; Lovering, A.M.; Webb, J.C. The role of microbial biofilms in prosthetic joint infections. Acta Orthop. 2015, 86, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Parthiban, C.; Varudharasu, D.; Shanmugam, M.; Gopal, P.; Ragunath, C.; Thomas, L.; Nitz, M.; Ramasubbu, N. Structural and functional analysis of de-N-acetylase PgaB from periodontopathogen Aggregatibacter actinomycetemcomitans. Mol. Oral Microbiol. 2017, 32, 324–340. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Attwood, T.K.; Babbitt, P.C.; Bateman, A.; Bork, P.; Bridge, A.J.; Chang, H.Y.; Dosztanyi, Z.; El-Gebali, S.; Fraser, M.; et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017, 45, D190–D199. [Google Scholar] [CrossRef] [PubMed]
- Tsalafouta, A.; Psylinakis, E.; Kapetaniou, E.G.; Kotsifaki, D.; Deli, A.; Roidis, A.; Bouriotis, V.; Kokkinidis, M. Purification, crystallization and preliminary X-ray analysis of the peptidoglycan N-acetylglucosamine deacetylase BC1960 from Bacillus cereus in the presence of its substrate (GlcNAc)6. Acta Crystallogr. Sect. F 2008, 64, 203–205. [Google Scholar] [CrossRef] [PubMed]
- Hernick, M.; Fierke, C.A. Zinc hydrolases: The mechanisms of zinc-dependent deacetylases. Arch. Biochem. Biophys. 2005, 433, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Asensio, J.L.; Ardá, A.; Cañada, F.J.; Jiménez-Barbero, J. Carbohydrate-aromatic interactions. Acc. Chem. Res. 2013, 46, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Santana, A.G.; Jiménez-Moreno, E.; Gómez, A.M.; Corzana, F.; González, C.; Jiménez-Oses, G.; Jiménez-Barbero, J.; Asensio, J.L. A dynamic combinatorial approach for the analysis of weak carbohydrate/aromatic complexes: Dissecting facial selectivity in CH/π stacking interactions. J. Am. Chem. Soc. 2013, 135, 3347–3350. [Google Scholar] [CrossRef] [PubMed]
- Planas, K.; Universitat Ramon Llull, Barcelona, Spain. Unpublished work. 2018.








| Subfamil (1) | Enzyme | Organism | PDB (Year) | Ref (2) | Polymer Substrates | COS Substrates (3) | Metal | PA (4) (on An) |
|---|---|---|---|---|---|---|---|---|
| Chitin DA | MrCDA | Mucor rouxii | -- | Chitin, chitosan | ≥DP3 | Zn2+ | Dn, Dn−1A | |
| ClCDA | Colletotrichum lindemuthianum | 2IW0 (2006) | [61] | Glycol-chitin | DP6>DP5>DP4>DP3>DP2 | Co2+ Zn2+ | Dn | |
| AnCDA | Aspergillus nidulans | 2Y8U (2012) | [12] | Glycol-chitin, chitin, CM-chitin, acetylxylan | DP2>DP3>DP4>DP5 | Co2+ | Dn | |
| PgtCDA | Puccinia graminis | -- | Glycol-chitin, colloidal chitin, chitosans | DP6>DP5>DP4 | n.r. (5) | AADn−2 | ||
| PesCDA | Pestolotiopsis sp. | -- | Colloidal chitin, chitosan DA10-60% | DP6-DP5-DP4 | n.r. | AADn−3A | ||
| PaCDA | Podospora anserina | -- | Glycol-chitin | ≥DP2 | Zn2+ | Dn | ||
| PcCDA | Pochonia chlamydosporia | -- | n.r. | DP5>DP4 | n.r. | ADDAn−3 | ||
| NodB | Sinorhizobium meliloti | -- | COS | DP5>DP2 (DP4, DP3) | Mn2+ Mg2+ | DAn−1 | ||
| VcCDA (COD) | Vibrio cholera | 4NY2 (2014) | [62] | COS | DP2>DP3>DP4>DP5>DP6 | Zn2+ | ADAn−2 | |
| VpCDA (COD) | Vibrio parahaemolyticus | 3WX7 (2014) | [63] | COS | DP2, DP3 | Zn2+ | n.r. | |
| ArCE4 | Arthrobacter sp. | 5LFZ (2017) | [64] | Chitin, chitosan, acetylxylan | DP5>DP6=DP4>DP3>>DP2 | Ni2+ (6) | A3D2 | |
| GlcNAc DA | SpPgdA | Streptococcus pneumoniae | 2C1G (2005) | [65] | GlcNAc DA on peptido-glycan | (GlcNAc)3 | Zn2+ | ADA |
| SmPgdA | Streptococcus mutants | 2W3Z (2008) | [66] | GlcNAc DA on peptidoglycan | DP6 | Zn2+ | n.r. | |
| BcPgd (BC1960) | Bacillus cereus | 4L1G (2014) | [67] | GlcNAc DA on peptido-glycan, glycol-chitin | DP6-DP5-DP4>>DP3>DP2 | Co2+ | Dn−1A | |
| ErPgd | Eubacterium rectale | 5JMU (2016) | GlcNAc deacetylase (annotated) | Zn2+ | ||||
| MurNAc DA | BsPdaA | Bacillus subtilis | 1W17 (2005) | [68] | MurNAc DA on peptido-glycan (Cwld digested) | No active on COS | Cd2+ (6) | |
| BaPda (BA0424) | Bacillus anthracis | 2J13 (2006) | [69] | MurNAc DA on peptido-glycan | n.r. | Zn2+ | ||
| PPda (unk) | BC0361 | Bacillus cereus | 4HD5 (2012) | [70] | Substrate unknown Putative GlcNAc DA | Zn2+ | ||
| BA0330 | Bacillus anthracis | 4V33 (2015) | [71] | Unknown. Not active on glycol-chitin, COS, pNPAc, synthetic muropeptide | Zn2+ | |||
| BA0150 | Bacillus anthracis | 4M1B (2014) | [72] | Presumably inactive (no metal coordination) | No metal | |||
| ECU11_0510 | Encephalitozoon cuniculi | 2VYO (2009) | [73] | Inactive (lack of Asp general base and His metal-binding) | No metal | |||
| AXE | SlAxeA | Streptomyces lividans | 2CC0 (2006) | [74] | Acetylxylan, glycol-chitin, chitosan | DP2-DP4-DP6 | Co2+ | DDD (A1D2) |
| CtAxeA | Clostridium thermocellum | 2C71 (2006) | [74] | 2-O-acetylxylan | No active on COS | Co2+ | ||
| β-1,6-GlcNAc DA | EcPgaB | Escherichia coli | 3VUS (2012) | [75,76] | Poly-β-1,6-GlcNAc de-N-acetylase | β-1,6-GlcNAc oligomers | Co2+ Ni2+ Zn2+ | |
| AdIcaB | Ammonifex degensii | 4WCJ (2014) | [77] | Poly-β-1,6-GlcNAc de-N-acetylase | β-1,6-GlcNAc oligomers | Ni2+ Co2+ Zn2+ | ||
| BbBpsB | Bordetella bronchiseptica | 5BU6 (2015) | [45] | Poly-β-1,6-GlcNAc de-N-acetylase | β-1,6-GlcNAc oligomers | Ni2+ Co2+ | ||
| AaPgaB | Aggregatibacter actinomycetemcomitans | 4U10 (2015) | Poly-β-1,6-GlcNAc de-N-acetylase | β-1,6-GlcNAc oligomers | Zn2+ |
| Enzyme | Uniprot AC | # aa FL 1 | CE4 (aa) 2 | Modular Structure |
|---|---|---|---|---|
| MrCDA | CDA_AMYRO | 421 | 151–349 |  |
| ClCDA | Q6DWK3_COLLN | 248 | 30–246 |  |
| AnCDA | B3VD85_EMEND | 249 | 44–245 |  |
| PgtCDA | E3K3D7_PUCGT | 269 | 38–236 |  |
| PesCDA | A0A1L3THR9_9PEZI | 298 | 27–236 |  |
| PaCDA | B2AAQ0_PODAN | 396 | 120–307 |  |
| PcCDA | -- | 455 | 107–303 |  |
| NodB | NODB_RHIME | 217 | 15–213 |  |
| VcCDA | Q9KSH6_VIBCH | 431 | 26–338 |  |
| VpCDA | A6P4T5_VIBPH | 427 | 28–326 |  |
| ArCE4 | A0A2C8C1T7_9MICC | 246 | 42–227 |  |
| SpPgdA | Q8DP63_STRR6 | 463 | 264–454 |  |
| SmPgdA | Q8DV82_STRMU | 311 | 103–308 |  |
| BcPgd BC1960 | B9J460_BACCQ | 275 | 68–266 |  |
| ErPgd | C4ZEZ9_AGARV | 496 | 290–482 |  |
| BsPdaA | PDAA_BACSU | 266 | 68–253 |  |
| BaPda | Q81Z49_BACAN | 260 | 45–255 |  |
| BC0361 | Q81IM3_BACCR | 360 | 195–360 |  |
| BA0330 | Q81ZD9_BACAN | 360 | 195–360 |  |
| BA0150 | Q81VP2_BACAN | 254 | 52–237 |  |
| ECU11_ | YB51_ENCCU | 254 | 26–210 |  |
| SlAxeA | Q54413_STRLI | 335 | 44–221 |  |
| CtAxe (XynA) | O87119_CLOTM | 683 | 477–655 |  |
| EcPgaB | PGAB_ECOLI | 672 | 65–349 |  |
| AdIcaB | C9RCK9_AMMDK | 280 | 67–280 |  |
| BbBpsB | A0A058YIS5_BORBO | 701 | 66–355 |  |
| AaPgaB | A5HJW8_AGGAC | 638 | 48–334 |  |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aragunde, H.; Biarnés, X.; Planas, A. Substrate Recognition and Specificity of Chitin Deacetylases and Related Family 4 Carbohydrate Esterases. Int. J. Mol. Sci. 2018, 19, 412. https://doi.org/10.3390/ijms19020412
Aragunde H, Biarnés X, Planas A. Substrate Recognition and Specificity of Chitin Deacetylases and Related Family 4 Carbohydrate Esterases. International Journal of Molecular Sciences. 2018; 19(2):412. https://doi.org/10.3390/ijms19020412
Chicago/Turabian StyleAragunde, Hugo, Xevi Biarnés, and Antoni Planas. 2018. "Substrate Recognition and Specificity of Chitin Deacetylases and Related Family 4 Carbohydrate Esterases" International Journal of Molecular Sciences 19, no. 2: 412. https://doi.org/10.3390/ijms19020412
APA StyleAragunde, H., Biarnés, X., & Planas, A. (2018). Substrate Recognition and Specificity of Chitin Deacetylases and Related Family 4 Carbohydrate Esterases. International Journal of Molecular Sciences, 19(2), 412. https://doi.org/10.3390/ijms19020412