Epigenetics and MicroRNAs in Cancer
Abstract
:1. Introduction
2. Epigenetic Alterations of miRNAs in Cancer
2.1. By DNA Methylation
2.2. By Histone Modifications
3. MiRNAs as Epigenetic Regulators
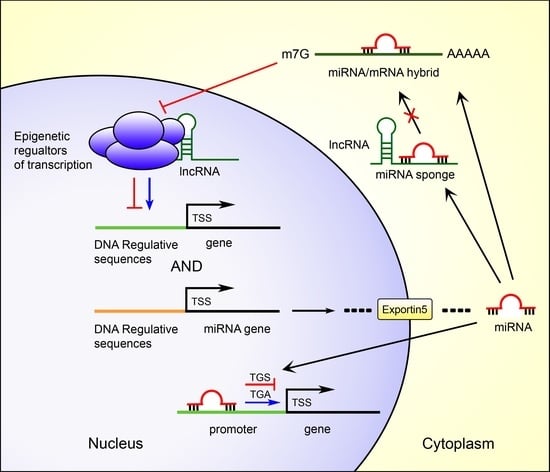
3.1. Post-Transcriptional Gene Silencing by miRNAs
3.2. miRNAs Regulate Gene Transcription
3.2.1. MiRNAs Transcriptional Gene Silencing (TGS)
3.2.2. MiRNAs Transcriptional Gene Activation (TGA)
4. Others
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Waddington, C.H. The epigenotype. Endeavour 1942, 1, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Holliday, R. The inheritance of epigenetic defects. Science 1987, 238, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Holliday, R. Epigenetics: An overview. Dev. Genet. 1994, 15, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Russo, V.E.A.; Martienssen, R.A.; Riggs, A.D. Epigenetic Mechanisms of Gene Regulation; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 1996. [Google Scholar]
- Wu, C.; Morris, J.R. Genes, genetics, and epigenetics: A correspondence. Science 2001, 293, 1103–1105. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Mersfelder, E.L.; Parthun, M.R. The tale beyond the tail: Histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 2006, 34, 2653–2662. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Allis, C.D. Rna meets chromatin. Genes Dev. 2005, 19, 1635–1655. [Google Scholar] [CrossRef] [PubMed]
- Murtha, M.; Esteller, M. Extraordinary cancer epigenomics: Thinking outside the classical coding and promoter box. Trends Cancer 2016, 2, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 629–656. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The epigenomics of cancer. Cell 2007, 128, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Easwaran, H.; Tsai, H.C.; Baylin, S.B. Cancer epigenetics: Tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell 2014, 54, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Wainwright, E.N.; Scaffidi, P. Epigenetics and cancer stem cells: Unleashing, hijacking, and restricting cellular plasticity. Trends Cancer 2017, 3, 372–386. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P.; Ohlsson, R.; Henikoff, S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006, 7, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Cairns, B.R. The logic of chromatin architecture and remodelling at promoters. Nature 2009, 461, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Comet, I.; Riising, E.M.; Leblanc, B.; Helin, K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer 2016, 16, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.G.; Roberts, C.W. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 2011, 11, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.Y.; Wade, P.A. Cancer biology and nurd: A multifaceted chromatin remodelling complex. Nat. Rev. Cancer 2011, 11, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Beck, D.B.; Oda, H.; Shen, S.S.; Reinberg, D. Pr-Set7 and H4K20me1: At the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012, 26, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Thiagalingam, S.; Cheng, K.H.; Lee, H.J.; Mineva, N.; Thiagalingam, A.; Ponte, J.F. Histone deacetylases: Unique players in shaping the epigenetic histone code. Ann. N. Y. Acad. Sci. 2003, 983, 84–100. [Google Scholar] [CrossRef] [PubMed]
- Saxena, A.; Carninci, P. Long non-coding rna modifies chromatin: Epigenetic silencing by long non-coding RNAs. Bioessays 2011, 33, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Forrest, M.E.; Khalil, A.M. Review: Regulation of the cancer epigenome by long non-coding RNAs. Cancer Lett. 2017, 407, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of microRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Lovat, F.; Valeri, N.; Croce, C.M. MicroRNAs in the pathogenesis of cancer. Semin. Oncol. 2011, 38, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Maruyama, R.; Yamamoto, E.; Kai, M. DNA methylation and microRNA dysregulation in cancer. Mol. Oncol. 2012, 6, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C.; et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3a and 3b. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Liu, S.; Fabbri, M.; Liu, Z.; Heaphy, C.E.; Callegari, E.; Schwind, S.; Pang, J.; Yu, J.; Muthusamy, N.; et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood 2009, 113, 6411–6418. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Stedman, W.; Yousef, M.; Renne, R.; Lieberman, P.M. Epigenetic regulation of kaposi’s sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J. Virol. 2010, 84, 2697–2706. [Google Scholar] [CrossRef] [PubMed]
- Braconi, C.; Huang, N.; Patel, T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 2010, 51, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The emt-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Liu, X.; Fu, H.; Sun, Y.; Wang, L.; Xu, G.; Wang, W.; Yu, Z.; Liu, C.; Li, P.; et al. miR-101 reverses hypomethylation of the PRDM16 promoter to disrupt mitochondrial function in astrocytoma cells. Oncotarget 2016, 7, 5007–5022. [Google Scholar] [CrossRef] [PubMed]
- Park, C.W.; Zeng, Y.; Zhang, X.; Subramanian, S.; Steer, C.J. Mature microRNAs identified in highly purified nuclei from HCT116 colon cancer cells. RNA Biol. 2010, 7, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.Y.; Ma, L.M.; Guo, Y.H.; Zhang, Y.C.; Zhou, H.; Shao, P.; Chen, Y.Q.; Qu, L.H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS ONE 2010, 5, e10563. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.J.; Bird, A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Saxonov, S.; Berg, P.; Brutlag, D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 2006, 103, 1412–1417. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.; Stresemann, C.; Brueckner, B.; Lyko, F. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle 2007, 6, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Liang, G.; Egger, G.; Friedman, J.M.; Chuang, J.C.; Coetzee, G.A.; Jones, P.A. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 2006, 9, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Lujambio, A.; Ropero, S.; Ballestar, E.; Fraga, M.F.; Cerrato, C.; Setien, F.; Casado, S.; Suarez-Gauthier, A.; Sanchez-Cespedes, M.; Git, A.; et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007, 67, 1424–1429. [Google Scholar] [CrossRef] [PubMed]
- Lujambio, A.; Calin, G.A.; Villanueva, A.; Ropero, S.; Sanchez-Cespedes, M.; Blanco, D.; Montuenga, L.M.; Rossi, S.; Nicoloso, M.S.; Faller, W.J.; et al. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA 2008, 105, 13556–13561. [Google Scholar] [CrossRef] [PubMed]
- Pronina, I.V.; Loginov, V.I.; Burdennyy, A.M.; Fridman, M.V.; Senchenko, V.N.; Kazubskaya, T.P.; Kushlinskii, N.E.; Dmitriev, A.A.; Braga, E.A. DNA methylation contributes to deregulation of 12 cancer-associated microRNAs and breast cancer progression. Gene 2017, 604, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Deatherage, D.E.; Rodriguez, B.A.; Liyanarachchi, S.; Weng, Y.I.; Zuo, T.; Liu, J.; Cheng, A.S.; Huang, T.H. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer Res. 2009, 69, 5936–5945. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, U.; Hasemeier, B.; Christgen, M.; Muller, M.; Romermann, D.; Langer, F.; Kreipe, H. Epigenetic inactivation of microRNA gene hsa-miR-9-1 in human breast cancer. J. Pathol. 2008, 214, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Katsaros, D.; Zhu, Y.; Hoffman, A.; Luca, S.; Marion, C.E.; Mu, L.; Risch, H.; Yu, H. Let-7a regulation of insulin-like growth factors in breast cancer. Breast Cancer Res. Treat. 2011, 126, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Biagioni, F.; Bossel Ben-Moshe, N.; Fontemaggi, G.; Canu, V.; Mori, F.; Antoniani, B.; Di Benedetto, A.; Santoro, R.; Germoni, S.; De Angelis, F.; et al. miR-10b*, a master inhibitor of the cell cycle, is down-regulated in human breast tumours. EMBO Mol. Med. 2012, 4, 1214–1229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, L.X.; Wu, Q.N.; Du, Z.M.; Chen, J.; Liao, D.Z.; Huang, M.Y.; Hou, J.H.; Wu, Q.L.; Zeng, M.S.; et al. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 2011, 71, 3552–3562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, P.; Sun, T.; Li, D.; Xu, X.; Rui, Y.; Li, C.; Chong, M.; Ibrahim, T.; Mercatali, L.; et al. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat. Cell Biol. 2013, 15, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Jiang, Y.; Yin, Y.; Li, Q.; He, J.; Jing, Y.; Qi, Y.T.; Xu, Q.; Li, W.; Lu, B.; et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 2013, 5, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhao, Y.; Liu, C.; Chen, X.; Qi, Y.; Jiang, Y.; Zou, C.; Zhang, X.; Liu, S.; Wang, X.; et al. Analysis of miR-195 and miR-497 expression, regulation and role in breast cancer. Clin. Cancer Res. 2011, 17, 1722–1730. [Google Scholar] [CrossRef] [PubMed]
- Castilla, M.A.; Diaz-Martin, J.; Sarrio, D.; Romero-Perez, L.; Lopez-Garcia, M.A.; Vieites, B.; Biscuola, M.; RamiRo-Fuentes, S.; Isacke, C.M.; Palacios, J. MicroRNA-200 family modulation in distinct breast cancer phenotypes. PLoS ONE 2012, 7, e47709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haga, C.L.; Phinney, D.G. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J. Biol. Chem. 2012, 287, 42695–42707. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, U. Aberrant DNA methylation of microRNA genes in human breast cancer—A critical appraisal. Cell Tissue Res. 2014, 356, 657–664. [Google Scholar] [CrossRef] [PubMed]
- He, D.X.; Gu, X.T.; Li, Y.R.; Jiang, L.; Jin, J.; Ma, X. Methylation-regulated miR-149 modulates chemoresistance by targeting glcnac N-deacetylase/N-sulfotransferase-1 in human breast cancer. FEBS J. 2014, 281, 4718–4730. [Google Scholar] [CrossRef] [PubMed]
- Omura, N.; Li, C.P.; Li, A.; Hong, S.M.; Walter, K.; Jimeno, A.; Hidalgo, M.; Goggins, M. Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol. Ther. 2008, 7, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chen, L.; Zhang, J.; Chen, H.; Fan, J.; Wang, K.; Luo, J.; Chen, Z.; Meng, Z.; Liu, L. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene 2014, 33, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Gu, Y.; Li, Z.; Cai, H.; Peng, Q.; Tu, M.; Kondo, Y.; Shinjo, K.; Zhu, Y.; Zhang, J.; et al. miR-615-5p is epigenetically inactivated and functions as a tumor suppressor in pancreatic ductal adenocarcinoma. Oncogene 2015, 34, 1629–1640. [Google Scholar] [CrossRef] [PubMed]
- Botla, S.K.; Savant, S.; Jandaghi, P.; Bauer, A.S.; Mucke, O.; Moskalev, E.A.; Neoptolemos, J.P.; Costello, E.; Greenhalf, W.; Scarpa, A.; et al. Early epigenetic downregulation of microRNA-192 expression promotes pancreatic cancer progression. Cancer Res. 2016, 76, 4149–4159. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.M.; Kang, E.J.; Kwon, H.M.; Bae, J.H.; Kang, K.; Ahuja, N.; Yang, K. Epigenetically altered miR-1247 functions as a tumor suppressor in pancreatic cancer. Oncotarget 2017, 8, 26600–26612. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Omura, N.; Hong, S.M.; Vincent, A.; Walter, K.; Griffith, M.; Borges, M.; Goggins, M. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010, 70, 5226–5237. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Yamamoto, E.; Nojima, M.; Kai, M.; Yamano, H.O.; Yoshikawa, K.; Kimura, T.; Kudo, T.; Harada, E.; Sugai, T.; et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis 2010, 31, 2066–2073. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Akiyama, Y.; Otsubo, T.; Shimada, S.; Yuasa, Y. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis 2010, 31, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.W.; Hu, L.Y.; Chen, T.W.; Li, S.C.; Ho, M.R.; Yu, S.Y.; Tu, Y.T.; Chen, W.S.; Lam, H.C. Emerging role of microRNAs in modulating endothelin-1 expression in gastric cancer. Oncol. Rep. 2015, 33, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.W.; Liao, Y.L.; Wu, C.W.; Hu, L.Y.; Li, S.C.; Chan, W.C.; Ho, M.R.; Lai, C.H.; Kao, H.W.; Fang, W.L.; et al. Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics 2011, 6, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.W.; Wu, C.W.; Hu, L.Y.; Li, S.C.; Liao, Y.L.; Lai, C.H.; Kao, H.W.; Fang, W.L.; Huang, K.H.; Chan, W.C.; et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int. J. Cancer 2011, 129, 2600–2610. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Zhang, Z.; Zou, D.; Wang, B.; Yan, Y.; Luo, M.; Dong, L.; Yin, H.; Gong, B.; Li, Z.; et al. microRNA-10a is down-regulated by DNA methylation and functions as a tumor suppressor in gastric cancer cells. PLoS ONE 2014, 9, e88057. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lei, H.; Luo, M.; Wang, Y.; Dong, L.; Ma, Y.; Liu, C.; Song, W.; Wang, F.; Zhang, J.; et al. DNA methylation downregulated miR-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer 2015, 18, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.; Shi, Z.; Liu, X.; Zhang, A.; Han, L.; Jiang, K.; Kang, C.; Zhang, Q. DNMT1 and EZH2 mediated methylation silences the microRNA-200b/a/429 gene and promotes tumor progression. Cancer Lett. 2015, 359, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Song, P.; Su, R.; Yang, G.; Dong, L.; Luo, M.; Wang, B.; Gong, B.; Liu, C.; Song, W.; et al. DNA methylation mediated down-regulating of microRNA-33b and its role in gastric cancer. Sci. Rep. 2016, 6, 18824. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Yoshida, T.; Enomoto, S.; Asada, K.; Tatematsu, M.; Ichinose, M.; Sugiyama, T.; Ushijima, T. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: Its possible involvement in the formation of epigenetic field defect. Int. J. Cancer 2009, 124, 2367–2374. [Google Scholar] [CrossRef] [PubMed]
- Datta, J.; Kutay, H.; Nasser, M.W.; Nuovo, G.J.; Wang, B.; Majumder, S.; Liu, C.G.; Volinia, S.; Croce, C.M.; Schmittgen, T.D.; et al. Methylation mediated silencing of microRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008, 68, 5049–5058. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cheng, J.; Zeng, Z.; Wang, Y.; Li, X.; Xie, Q.; Jia, J.; Yan, Y.; Guo, Z.; Gao, J.; et al. Comprehensive profiling of novel microRNA-9 targets and a tumor suppressor role of microRNA-9 via targeting IGF2BP1 in hepatocellular carcinoma. Oncotarget 2015, 6, 42040–42052. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Liu, J.; Chen, J.; Dong, J.; Ma, H.; Liu, Y.; Hu, Z. Methylation-associated silencing of microRNA-34b in hepatocellular carcinoma cancer. Gene 2014, 543, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Furuta, M.; Kozaki, K.I.; Tanaka, S.; Arii, S.; Imoto, I.; Inazawa, J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis 2010, 31, 766–776. [Google Scholar] [CrossRef] [PubMed]
- He, X.X.; Kuang, S.Z.; Liao, J.Z.; Xu, C.R.; Chang, Y.; Wu, Y.L.; Gong, J.; Tian, D.A.; Guo, A.Y.; Lin, J.S. The regulation of microRNA expression by DNA methylation in hepatocellular carcinoma. Mol. Biosyst. 2015, 11, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Veronese, A.; Visone, R.; Consiglio, J.; Acunzo, M.; Lupini, L.; Kim, T.; Ferracin, M.; Lovat, F.; Miotto, E.; Balatti, V.; et al. Mutated beta-catenin evades a microRNA-dependent regulatory loop. Proc. Natl. Acad. Sci. USA 2011, 108, 4840–4845. [Google Scholar] [CrossRef] [PubMed]
- Callegari, E.; Elamin, B.K.; Giannone, F.; Milazzo, M.; Altavilla, G.; Fornari, F.; Giacomelli, L.; D’Abundo, L.; Ferracin, M.; Bassi, C.; et al. Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology 2012, 56, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Milazzo, M.; Galassi, M.; Callegari, E.; Veronese, A.; Miyaaki, H.; Sabbioni, S.; Mantovani, V.; Marasco, E.; Chieco, P.; et al. P53/mdm2 feedback loop sustains miR-221 expression and dictates the response to anticancer treatments in hepatocellular carcinoma. Mol. Cancer Res. MCR 2014, 12, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Nojima, M.; Matsui, T.; Tamori, A.; Kubo, S.; Shirabe, K.; Kimura, K.; Shimada, M.; Utsunomiya, T.; Kondo, Y.; Iio, E.; et al. Global, cancer-specific microRNA cluster hypomethylation was functionally associated with the development of non-b non-c hepatocellular carcinoma. Mol. Cancer 2016, 15, 31. [Google Scholar] [CrossRef] [PubMed]
- Selcuklu, S.D.; Donoghue, M.T.; Rehmet, K.; de Souza Gomes, M.; Fort, A.; Kovvuru, P.; Muniyappa, M.K.; Kerin, M.J.; Enright, A.J.; Spillane, C. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J. Biol. Chem. 2012, 287, 29516–29528. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kelnar, K.; Liu, B.; Chen, X.; Calhoun-Davis, T.; Li, H.; Patrawala, L.; Yan, H.; Jeter, C.; Honorio, S.; et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 2011, 17, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Park, E.Y.; Chang, E.; Lee, E.J.; Lee, H.W.; Kang, H.G.; Chun, K.H.; Woo, Y.M.; Kong, H.K.; Ko, J.Y.; Suzuki, H.; et al. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 2014, 74, 7573–7582. [Google Scholar] [CrossRef] [PubMed]
- Bu, P.; Chen, K.Y.; Chen, J.H.; Wang, L.; Walters, J.; Shin, Y.J.; Goerger, J.P.; Sun, J.; Witherspoon, M.; Rakhilin, N.; et al. A microRNA miR-34a-regulated bimodal switch targets notch in colon cancer stem cells. Cell Stem Cell 2013, 12, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Cama, A.; Verginelli, F.; Lotti, L.V.; Napolitano, F.; Morgano, A.; D’Orazio, A.; Vacca, M.; Perconti, S.; Pepe, F.; Romani, F.; et al. Integrative genetic, epigenetic and pathological analysis of paraganglioma reveals complex dysregulation of notch signaling. Acta Neuropathol. 2013, 126, 575–594. [Google Scholar] [CrossRef] [PubMed]
- Hermeking, H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010, 17, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.H.; Sun, M.; Nie, F.Q.; Ge, Y.B.; Zhang, E.B.; Yin, D.D.; Kong, R.; Xia, R.; Lu, K.H.; Li, J.H.; et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer 2014, 13. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Song, L.; Zeng, S.; Zhang, L. MALAT1-miR-124-RBG2 axis is involved in growth and invasion of HR-HPV-positive cervical cancer cells. Tumour Biol. 2016, 37, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Zhao, N.; Zha, G.; Wang, H.; Tong, Q.; Xin, S. LncRNA HOXA11-AS exerts oncogenic functions by repressing p21 and miR-124 in uveal melanoma. DNA Cell Biol. 2017, 36, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, Z.; Zhou, Z.; Liu, R. Linc-ROR confers gemcitabine resistance to pancreatic cancer cells via inducing autophagy and modulating the miR-124/PTBP1/PKM2 axis. Cancer Chemother. Pharmacol. 2016, 78, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding rna networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Nie, F.; Wang, Y.; Zhang, Z.; Hou, J.; He, D.; Xie, M.; Xu, L.; De, W.; Wang, Z.; et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016, 76, 6299–6310. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ror induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lu, L.; Feng, B.; Zhang, K.; Han, S.; Hou, D.; Chen, L.; Chu, X.; Wang, R. The lincRNA-ROR/miR-145 axis promotes invasion and metastasis in hepatocellular carcinoma via induction of epithelial-mesenchymal transition by targeting ZEB2. Sci. Rep. 2017, 7, 4637. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, Y.; Shen, H.; Li, H.; Long, L.; Hui, L.; Xu, W. miR-137 inhibits the proliferation of lung cancer cells by targeting Cdc42 and Cdk6. FEBS Lett. 2013, 587, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Xiao, D.; Zhao, Z.; Ren, P.; Li, C.; Hu, Y.; Shi, J.; Su, H.; Wang, L.; Liu, H.; et al. Epigenetic silencing of microRNA-137 enhances ASCT2 expression and tumor glutamine metabolism. Oncogenesis 2017, 6, e356. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zheng, G.; Gu, Z.; Guo, Z. miR-137 inhibits proliferation and angiogenesis of human glioblastoma cells by targeting EZH2. J. Neurooncol. 2015, 122, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Ng, S.S.; Jiang, S.; Cheung, W.K.; Sze, J.; Bian, X.W.; Kung, H.F.; Lin, M.C. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem. Biophys. Res. Commun. 2010, 391, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Park, S.M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the e-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Veronese, A.; Pichiorri, F.; Lee, T.J.; Jeon, Y.J.; Volinia, S.; Pineau, P.; Marchio, A.; Palatini, J.; Suh, S.S.; et al. P53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J. Exp. Med. 2011, 208, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Tang, J.; Lei, H.; Cai, P.; Zhu, H.; Li, B.; Xu, X.; Xia, Y.; Tang, W. Decreased miR-200a/141 suppress cell migration and proliferation by targeting pten in hirschsprung’s disease. Cell Physiol. Biochem. 2014, 34, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Wagner, E.; Roidl, A. The proto-oncogene kras is targeted by miR-200c. Oncotarget 2014, 5, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.Y.; Wright, J.A.; Attema, J.L.; Gregory, P.A.; Bert, A.G.; Smith, E.; Thomas, D.; Lopez, A.F.; Drew, P.A.; Khew-Goodall, Y.; et al. Epigenetic modulation of the miR-200 family is associated with transition to a breast cancer stem-cell-like state. J. Cell Sci. 2013, 126, 2256–2266. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T. Re-set-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001, 11, 266–273. [Google Scholar] [CrossRef]
- Scott, G.K.; Mattie, M.D.; Berger, C.E.; Benz, S.C.; Benz, C.C. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 2006, 66, 1277–1281. [Google Scholar] [CrossRef] [PubMed]
- Sampath, D.; Liu, C.; Vasan, K.; Sulda, M.; Puduvalli, V.K.; Wierda, W.G.; Keating, M.J. Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood 2012, 119, 1162–1172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, X.; Lin, J.; Lwin, T.; Wright, G.; Moscinski, L.C.; Dalton, W.S.; Seto, E.; Wright, K.; Sotomayor, E.; et al. Myc represses miR-15a/miR-16-1 expression through recruitment of HDAC3 in mantle cell and other non-hodgkin b-cell lymphomas. Oncogene 2012, 31, 3002–3008. [Google Scholar] [CrossRef] [PubMed]
- Mertens, D.; Wolf, S.; Tschuch, C.; Mund, C.; Kienle, D.; Ohl, S.; Schroeter, P.; Lyko, F.; Dohner, H.; Stilgenbauer, S.; et al. Allelic silencing at the tumor-suppressor locus 13q14.3 suggests an epigenetic tumor-suppressor mechanism. Proc. Natl. Acad. Sci. USA 2006, 103, 7741–7746. [Google Scholar] [CrossRef] [PubMed]
- Veronese, A.; Pepe, F.; Chiacchia, J.; Pagotto, S.; Lanuti, P.; Veschi, S.; Di Marco, M.; D’Argenio, A.; Innocenti, I.; Vannata, B.; et al. Allele-specific loss and transcription of the miR-15a/16-1 cluster in chronic lymphocytic leukemia. Leukemia 2015, 29, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Wang, R.H.; Akagi, K.; Kim, K.A.; Martin, B.K.; Cavallone, L.; Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab); Haines, D.C.; Basik, M.; Mai, P.; et al. Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat. Med. 2011, 17, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Sun, M.; Zhou, S.; Guo, B. Class I HDAC inhibitor mocetinostat induces apoptosis by activation of miR-31 expression and suppression of E2F6. Cell Death Discov. 2016, 2, 16036. [Google Scholar] [CrossRef] [PubMed]
- Buurman, R.; Gurlevik, E.; Schaffer, V.; Eilers, M.; Sandbothe, M.; Kreipe, H.; Wilkens, L.; Schlegelberger, B.; Kuhnel, F.; Skawran, B. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology 2012, 143, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Toh, H.C.; Chow, P.; Chung, A.Y.; Meyers, D.J.; Cole, P.A.; Ooi, L.L.; Lee, C.G. MicroRNA-224 is up-regulated in hepatocellular carcinoma through epigenetic mechanisms. FASEB J. 2012, 26, 3032–3041. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Feng, M.; Jiang, X.; Wu, Z.; Li, Z.; Aau, M.; Yu, Q. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate PRB-E2F1 activity through a feedback loop by targeting CDK6 and CDC25a. Genes Dev. 2009, 23, 2388–2393. [Google Scholar] [CrossRef] [PubMed]
- Au, S.L.; Wong, C.C.; Lee, J.M.; Fan, D.N.; Tsang, F.H.; Ng, I.O.; Wong, C.M. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology 2012, 56, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Mani, R.S.; Ateeq, B.; Dhanasekaran, S.M.; Asangani, I.A.; Prensner, J.R.; Kim, J.H.; Brenner, J.C.; Jing, X.; Cao, X.; et al. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell 2011, 20, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Padi, S.K.; Tindall, D.J.; Guo, B. Polycomb protein EZH2 suppresses apoptosis by silencing the proapoptotic miR-31. Cell Death Dis. 2014, 5, e1486. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.S.; Leung, C.M.; Pan, H.W.; Hu, L.Y.; Li, S.C.; Ho, M.R.; Tsai, K.W. Silencing of miR-1-1 and miR-133a-2 cluster expression by DNA hypermethylation in colorectal cancer. Oncol. Rep. 2012, 28, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Nasser, M.W.; Datta, J.; Nuovo, G.; Kutay, H.; Motiwala, T.; Majumder, S.; Wang, B.; Suster, S.; Jacob, S.T.; Ghoshal, K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J. Biol. Chem. 2008, 283, 33394–33405. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pan, Q.; Wan, X.; Mao, Y.; Lu, W.; Xie, X.; Cheng, X. Methylation-associated has-miR-9 deregulation in paclitaxel-resistant epithelial ovarian carcinoma. BMC Cancer 2015, 15, 509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, L.Q.; Wong, K.Y.; Li, Z.Y.; Chim, C.S. Infrequent DNA methylation of miR-9-1 and miR-9-3 in multiple myeloma. J. Clin. Pathol. 2015, 68, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, M.A.; Gu, J.; Lin, J.; Ye, Y.; Tan, W.; Tamboli, P.; Wood, C.G.; Wu, X. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene 2010, 29, 5724–5728. [Google Scholar] [CrossRef] [PubMed]
- Kohler, C.U.; Bryk, O.; Meier, S.; Lang, K.; Rozynek, P.; Bruning, T.; Kafferlein, H.U. Analyses in human urothelial cells identify methylation of miR-152, miR-200b and miR-10a genes as candidate bladder cancer biomarkers. Biochem. Biophys. Res. Commun. 2013, 438, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wang, S.; Zhang, Y.J.; Kappil, M.A.; Wu, H.C.; Kibriya, M.G.; Wang, Q.; Jasmine, F.; Ahsan, H.; Lee, P.H.; et al. Genome-wide aberrant DNA methylation of microRNA host genes in hepatocellular carcinoma. Epigenetics 2012, 7, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, K.J.; Cobiac, L.; Le Leu, R.K.; Van der Hoek, M.B.; Michael, M.Z. Histone deacetylase inhibition in colorectal cancer cells reveals competing roles for members of the oncogenic miR-17-92 cluster. Mol. Carcinog. 2013, 52, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.G.; Alder, H.; et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, A.; Kontos, C.K.; Boni, T.; Bantounas, I.; Siakouli, D.; Kosmidou, V.; Vlassi, M.; Spyridakis, Y.; Tsipras, I.; Zografos, G.; et al. Epigenetic regulation of miR-21 in colorectal cancer: ITGB4 as a novel miR-21 target and a three-gene network (miR-21-ITGBETA4-PDCD4) as predictor of metastatic tumor potential. Epigenetics 2014, 9, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Hulf, T.; Sibbritt, T.; Wiklund, E.D.; Bert, S.; Strbenac, D.; Statham, A.L.; Robinson, M.D.; Clark, S.J. Discovery pipeline for epigenetically deregulated miRnas in cancer: Integration of primary miRNA transcription. BMC Genom. 2011, 12, 54. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, R.; Claret, F.X.; Yang, H. Involvement of microRNA-24 and DNA methylation in resistance of nasopharyngeal carcinoma to ionizing radiation. Mol. Cancer Ther. 2014, 13, 3163–3174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, X.; Fiskus, W.; Lin, J.; Lwin, T.; Rao, R.; Zhang, Y.; Chan, J.C.; Fu, K.; Marquez, V.E.; et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 AND EZH2 as a therapeutic target of histone modification in aggressive B-cell lymphomas. Cancer Cell 2012, 22, 506–523. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wu, L.C.; Pang, J.; Santhanam, R.; Schwind, S.; Wu, Y.Z.; Hickey, C.J.; Yu, J.; Becker, H.; Maharry, K.; et al. Sp1/NFκB/HDAC/miR-29b regulatory network in kit-driven myeloid leukemia. Cancer Cell 2010, 17, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Dimri, M.; Dimri, G.P. MicroRNA-31 is a transcriptional target of histone deacetylase inhibitors and a regulator of cellular senescence. J. Biol. Chem. 2015, 290, 10555–10567. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.C.; Chiu, Y.L.; Banerjee, S.; Park, K.; Mosquera, J.M.; Giannopoulou, E.; Alves, P.; Tewari, A.K.; Gerstein, M.B.; Beltran, H.; et al. Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res. 2013, 73, 1232–1244. [Google Scholar] [CrossRef] [PubMed]
- Asangani, I.A.; Harms, P.W.; Dodson, L.; Pandhi, M.; Kunju, L.P.; Maher, C.A.; Fullen, D.R.; Johnson, T.M.; Giordano, T.J.; Palanisamy, N.; et al. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget 2012, 3, 1011–1025. [Google Scholar] [CrossRef] [PubMed]
- Augoff, K.; McCue, B.; Plow, E.F.; Sossey-Alaoui, K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol. Cancer 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, N.; Li, X.; Padi, S.K.; Zhang, Q.; Tang, M.S.; Guo, B. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. 2010, 1, e105. [Google Scholar] [CrossRef] [PubMed]
- Lodygin, D.; Tarasov, V.; Epanchintsev, A.; Berking, C.; Knyazeva, T.; Korner, H.; Knyazev, P.; Diebold, J.; Hermeking, H. Inactivation of miR-34a by aberrant cpg methylation in multiple types of cancer. Cell Cycle 2008, 7, 2591–2600. [Google Scholar] [CrossRef] [PubMed]
- Vogt, M.; Munding, J.; Gruner, M.; Liffers, S.T.; Verdoodt, B.; Hauk, J.; Steinstraesser, L.; Tannapfel, A.; Hermeking, H. Frequent concomitant inactivation of miR-34a and miR-34b/c by cpg methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011, 458, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Lee, W.K.; Lee, E.B.; Son, J.W.; Kim, D.S.; Park, J.Y. Combined effect of metastasis-related microRNA, miR-34 and miR-124 family, methylation on prognosis of non-small-cell lung cancer. Clin. Lung Cancer 2017, 18, e13–e20. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, T.; Soh, J.; Toyooka, S.; Aoe, K.; Fujimoto, N.; Hashida, S.; Maki, Y.; Tanaka, N.; Shien, K.; Furukawa, M.; et al. The degree of microRNA-34b/c methylation in serum-circulating DNA is associated with malignant pleural mesothelioma. Lung Cancer 2013, 82, 485–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parodi, F.; Carosio, R.; Ragusa, M.; Di Pietro, C.; Maugeri, M.; Barbagallo, D.; Sallustio, F.; Allemanni, G.; Pistillo, M.P.; Casciano, I.; et al. Epigenetic dysregulation in neuroblastoma: A tale of miRnas and DNA methylation. Biochim. Biophys. Acta 2016, 1859, 1502–1514. [Google Scholar] [CrossRef] [PubMed]
- Kozaki, K.; Imoto, I.; Mogi, S.; Omura, K.; Inazawa, J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008, 68, 2094–2105. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Lotterman, C.; Karikari, C.; Omura, N.; Feldmann, G.; Habbe, N.; Goggins, M.G.; Mendell, J.T.; Maitra, A. Epigenetic silencing of microRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology 2009, 9, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Chang, Y.; Guo, Y.; Wang, N.; Cui, J.; Gao, W.Q. Regulation and methylation of tumor suppressor miR-124 by androgen receptor in prostate cancer cells. PLoS ONE 2015, 10, e0116197. [Google Scholar] [CrossRef] [PubMed]
- Agirre, X.; Vilas-Zornoza, A.; Jimenez-Velasco, A.; Martin-Subero, J.I.; Cordeu, L.; Garate, L.; San Jose-Eneriz, E.; Abizanda, G.; Rodriguez-Otero, P.; Fortes, P.; et al. Epigenetic silencing of the tumor suppressor microRNA hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009, 69, 4443–4453. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, K.; Peters, I.; Dubrowinskaja, N.; Hennenlotter, J.; Abbas, M.; Scherer, R.; Tezval, H.; Merseburger, A.S.; Stenzl, A.; Kuczyk, M.A.; et al. Hsa-miR-124-3 CPG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br. J. Cancer 2013, 108, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Wilting, S.M.; van Boerdonk, R.A.; Henken, F.E.; Meijer, C.J.; Diosdado, B.; Meijer, G.A.; le Sage, C.; Agami, R.; Snijders, P.J.; Steenbergen, R.D. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol. Cancer 2010, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Silber, J.; Lim, D.A.; Petritsch, C.; Persson, A.I.; Maunakea, A.K.; Yu, M.; Vandenberg, S.R.; Ginzinger, D.G.; James, C.D.; Costello, J.F.; et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.; Kakar, S.; Kim, Y.S. MicroRNA-124a and microRNA-34b/c are frequently methylated in all histological types of colorectal cancer and polyps, and in the adjacent normal mucosa. Oncol. Lett. 2011, 2, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Alpini, G.; Glaser, S.S.; Zhang, J.P.; Francis, H.; Han, Y.; Gong, J.; Stokes, A.; Francis, T.; Hughart, N.; Hubble, L.; et al. Regulation of placenta growth factor by microRNA-125b in hepatocellular cancer. J. Hepatol. 2011, 55, 1339–1345. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.; Trapani, D.; Ravn, J.; Sorensen, J.B.; Andersen, C.B.; Grauslund, M.; Santoni-Rugiu, E. Methylation-associated silencing of microRNA-126 and its host gene EGFl7 in malignant pleural mesothelioma. Anticancer Res. 2015, 35, 6223–6229. [Google Scholar] [PubMed]
- Zhang, Y.; Wang, X.; Xu, B.; Wang, B.; Wang, Z.; Liang, Y.; Zhou, J.; Hu, J.; Jiang, B. Epigenetic silencing of miR-126 contributes to tumor invasion and angiogenesis in colorectal cancer. Oncol. Rep. 2013, 30, 1976–1984. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Friedman, J.M.; Chihara, Y.; Egger, G.; Chuang, J.C.; Liang, G. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem. Biophys. Res. Commun. 2009, 379, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Emoto, N.; Hamano, E.; Sunohara, M.; Kawakami, M.; Kage, H.; Kitano, K.; Nakajima, J.; Goto, A.; Fukayama, M.; et al. Genome structure-based screening identified epigenetically silenced microRNA associated with invasiveness in non-small-cell lung cancer. Int. J. Cancer 2012, 130, 2580–2590. [Google Scholar] [CrossRef] [PubMed]
- Wotschofsky, Z.; Liep, J.; Meyer, H.A.; Jung, M.; Wagner, I.; Disch, A.C.; Schaser, K.D.; Melcher, I.; Kilic, E.; Busch, J.; et al. Identification of metastamiRs as metastasis-associated microRNAs in clear cell renal cell carcinomas. Int. J. Biol. Sci. 2012, 8, 1363–1374. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, L.; Zhang, T.; Hao, M.; Zhang, X.; Zhang, J.; Xie, Q.; Wang, Y.; Guo, M.; Zhuang, H.; et al. Methylation-mediated repression of microRNA 129-2 enhances oncogenic SOX4 expression in HCC. Liver Int. 2013, 33, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.W.; Liu, J.C.; Deatherage, D.E.; Luo, J.; Mutch, D.G.; Goodfellow, P.J.; Miller, D.S.; Huang, T.H. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009, 69, 9038–9046. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Pan, S.; Qi, S.; Lin, X.; Cheng, S. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem. Biophys. Res. Commun. 2010, 394, 1047–1052. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.Y.; Yim, R.L.; Kwong, Y.L.; Leung, C.Y.; Hui, P.K.; Cheung, F.; Liang, R.; Jin, D.Y.; Chim, C.S. Epigenetic inactivation of the miR129-2 in hematological malignancies. J. Hematol. Oncol. 2013, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Bandres, E.; Agirre, X.; Bitarte, N.; RamiRez, N.; Zarate, R.; Roman-Gomez, J.; Prosper, F.; Garcia-Foncillas, J. Epigenetic regulation of microRNA expression in colorectal cancer. Int. J. Cancer 2009, 125, 2737–2743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hao, J.; Xie, F.; Hu, X.; Liu, C.; Tong, J.; Zhou, J.; Wu, J.; Shao, C. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis 2011, 32, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Formosa, A.; Lena, A.M.; Markert, E.K.; Cortelli, S.; Miano, R.; Mauriello, A.; Croce, N.; Vandesompele, J.; Mestdagh, P.; Finazzi-Agro, E.; et al. DNA methylation silences miR-132 in prostate cancer. Oncogene 2013, 32, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.V.; Zhou, J.; Lin, C.; Hu, G.; Yi, L.U.; Du, J.; Gao, K.; Li, X. DNA methylation is involved in the aberrant expression of miR-133b in colorectal cancer cells. Oncol. Lett. 2015, 10, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Zhang, S.; Deng, S.; An, G.; Qin, X.; Li, F.; Xu, Y.; Hao, M.; Yang, Y.; Zhou, W.; et al. Epigenetic silencing of miR-137 induces drug resistance and chromosomal instability by targeting AURKA in multiple myeloma. Leukemia 2017, 31, 1123–1135. [Google Scholar] [CrossRef] [PubMed]
- Langevin, S.M.; Stone, R.A.; Bunker, C.H.; Lyons-Weiler, M.A.; LaFramboise, W.A.; Kelly, L.; Seethala, R.R.; Grandis, J.R.; Sobol, R.W.; Taioli, E. MicroRNA-137 promoter methylation is associated with poorer overall survival in patients with squamous cell carcinoma of the head and neck. Cancer 2011, 117, 1454–1462. [Google Scholar] [CrossRef] [PubMed]
- Steponaitiene, R.; Kupcinskas, J.; Langner, C.; Balaguer, F.; Venclauskas, L.; Pauzas, H.; Tamelis, A.; Skieceviciene, J.; Kupcinskas, L.; Malfertheiner, P.; et al. Epigenetic silencing of miR-137 is a frequent event in gastric carcinogenesis. Mol. Carcinog. 2016, 55, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Balaguer, F.; Link, A.; Lozano, J.J.; Cuatrecasas, M.; Nagasaka, T.; Boland, C.R.; Goel, A. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010, 70, 6609–6618. [Google Scholar] [CrossRef] [PubMed]
- Daniunaite, K.; Dubikaityte, M.; Gibas, P.; Bakavicius, A.; Rimantas Lazutka, J.; Ulys, A.; Jankevicius, F.; Jarmalaite, S. Clinical significance of miRna host gene promoter methylation in prostate cancer. Hum. Mol. Genet. 2017, 26, 2451–2461. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Amano, Y.; Ishikawa, R.; Sunohara, M.; Kage, H.; Ichinose, J.; Sano, A.; Nakajima, J.; Fukayama, M.; Yatomi, Y.; et al. Histone methylation-mediated silencing of miR-139 enhances invasion of non-small-cell lung cancer. Cancer Med. 2015, 4, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.C.; Wong, C.M.; Tung, E.K.; Au, S.L.; Lee, J.M.; Poon, R.T.; Man, K.; Ng, I.O. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology 2011, 140, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.M.; O'Neill, K.M.; McKenna, M.M.; Walsh, C.P.; McKenna, D.J. Regulation of miR-200c and miR-141 by methylation in prostate cancer. Prostate 2016, 76, 1146–1159. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Zheng, D.; Li, J.; Li, Y.; Gao, L.; Wang, L.; Yu, L. Methylation-mediated repression of microRNA-143 enhances MLL-AF4 oncogene expression. Oncogene 2012, 31, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Chen, Q.; Wang, J.; Mao, Q.; Dong, G.; Shi, R.; Zheng, Y.; Xu, L.; Jiang, F. DNA methylation mediated silencing of microRNA-145 is a potential prognostic marker in patients with lung adenocarcinoma. Sci. Rep. 2015, 5, 16901. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Shen, N.; Weng, Y.; Li, K.; Hu, L.; Liao, H.; An, J.; Liu, L.; Lao, S.; Cai, S. Low miR-145 silenced by DNA methylation promotes NSCLC cell proliferation, migration and invasion by targeting mucin 1. Cancer Biol. Ther. 2015, 16, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.O.; Chen, Y.; Zaman, M.S.; Hirata, H.; Yamamura, S.; Shahryari, V.; Liu, J.; Tabatabai, Z.L.; Kakar, S.; Deng, G.; et al. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis 2011, 32, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Zaman, M.S.; Chen, Y.; Deng, G.; Shahryari, V.; Suh, S.O.; Saini, S.; Majid, S.; Liu, J.; Khatri, G.; Tanaka, Y.; et al. The functional significance of microRNA-145 in prostate cancer. Br. J. Cancer 2010, 103, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Tsuruta, T.; Kozaki, K.; Uesugi, A.; Furuta, M.; Hirasawa, A.; Imoto, I.; Susumu, N.; Aoki, D.; Inazawa, J. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. 2011, 71, 6450–6462. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, D.; Deb, M.; Rath, S.K.; Kar, S.; Parbin, S.; Pradhan, N.; Patra, S.K. DNA methylation and not H3K4 trimethylation dictates the expression status of miR-152 gene which inhibits migration of breast cancer cells via DNMT1/CDH1 loop. Exp. Cell Res. 2016, 346, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Ortega, E.; Arzate-Mejia, R.; Perez-Molina, R.; Gonzalez-Buendia, E.; Meier, K.; Guerrero, G.; Recillas-Targa, F. Epigenetic silencing of miR-181c by DNA methylation in glioblastoma cell lines. BMC Cancer 2016, 16, 226. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Cui, Y.; Wang, W.; Gu, J.; Guo, S.; Ma, K.; Luo, X. Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-miR-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma. Neoplasia 2011, 13, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; He, Y.; Zhang, H.; Fei, Q.; Niu, D.; Wang, D.; Ding, X.; Xu, H.; Chen, X.; Zhu, J. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J. Biol. Chem. 2012, 287, 5639–5649. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Deng, H.; Li, Y.; Zhang, C.; Liu, X.; Liu, Q.; Zhang, D.; Wang, L.; Pu, Y.; Zhang, H.; et al. The DNA methylation-regulated miR-193a-3p dictates the multi-chemoresistance of bladder cancer via repression of SRSF2/PLAU/HIC2 expression. Cell Death Dis. 2014, 5, e1402. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.N.; Lin, J.; Li, Y.H.; Gao, L.; Wang, X.R.; Wang, W.; Kang, H.Y.; Yan, G.T.; Wang, L.L.; Yu, L. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene 2011, 30, 3416–3428. [Google Scholar] [CrossRef] [PubMed]
- Torres-Ferreira, J.; Ramalho-Carvalho, J.; Gomez, A.; Menezes, F.D.; Freitas, R.; Oliveira, J.; Antunes, L.; Bento, M.J.; Esteller, M.; Henrique, R.; et al. miR-193b promoter methylation accurately detects prostate cancer in urine sediments and miR-34b/c or miR-129-2 promoter methylation define subsets of clinically aggressive tumors. Mol. Cancer 2017, 16, 26. [Google Scholar] [CrossRef] [PubMed]
- Rauhala, H.E.; Jalava, S.E.; Isotalo, J.; Bracken, H.; Lehmusvaara, S.; Tammela, T.L.; Oja, H.; Visakorpi, T. miR-193b is an epigenetically regulated putative tumor suppressor in prostate cancer. Int. J. Cancer 2010, 127, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.W.; Hu, L.Y.; Wu, C.W.; Li, S.C.; Lai, C.H.; Kao, H.W.; Fang, W.L.; Lin, W.C. Epigenetic regulation of miR-196b expression in gastric cancer. Genes Chromosom. Cancer 2010, 49, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Cheung, H.H.; Davis, A.J.; Lee, T.L.; Pang, A.L.; Nagrani, S.; Rennert, O.M.; Chan, W.Y. Methylation of an intronic region regulates miR-199a in testicular tumor malignancy. Oncogene 2011, 30, 3404–3415. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhao, F.; Hui, L.; Li, X.; Zhang, D.; Lin, W.; Chen, Z.; Ning, Y. Suppressing miR-199a-3p by promoter methylation contributes to tumor aggressiveness and cisplatin resistance of ovarian cancer through promoting DDR1 expression. J. Ovarian Res. 2017, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.R.; Sun, H.; Zhang, R.; Yu, X.H.; Shi, X.D.; Zhu, M.S.; Zeng, H.; Yan, L.X.; Xu, L.B.; Liu, C. Methylation-associated silencing of miR-200b facilitates human hepatocellular carcinoma progression by directly targeting BMI1. Oncotarget 2016, 7, 18684–18693. [Google Scholar] [CrossRef] [PubMed]
- Neves, R.; Scheel, C.; Weinhold, S.; Honisch, E.; Iwaniuk, K.M.; Trompeter, H.I.; Niederacher, D.; Wernet, P.; Santourlidis, S.; Uhrberg, M. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res. Notes 2010, 3, 219. [Google Scholar] [CrossRef] [PubMed]
- Ceppi, P.; Mudduluru, G.; Kumarswamy, R.; Rapa, I.; Scagliotti, G.V.; Papotti, M.; Allgayer, H. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol. Cancer Res. MCR 2010, 8, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Davalos, V.; Moutinho, C.; Villanueva, A.; Boque, R.; Silva, P.; Carneiro, F.; Esteller, M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 2012, 31, 2062–2074. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.W.; Kuo, C.T.; Chen, J.H.; Goodfellow, P.J.; Huang, T.H.; Rader, J.S.; Uyar, D.S. Hypermethylation of miR-203 in endometrial carcinomas. Gynecol. Oncol. 2014, 133, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Chim, C.S.; Wong, K.Y.; Leung, C.Y.; Chung, L.P.; Hui, P.K.; Chan, S.Y.; Yu, L. Epigenetic inactivation of the hsa-miR-203 in haematological malignancies. J. Cell Mol. Med. 2011, 15, 2760–2767. [Google Scholar] [CrossRef] [PubMed]
- Wiklund, E.D.; Bramsen, J.B.; Hulf, T.; Dyrskjot, L.; Ramanathan, R.; Hansen, T.B.; Villadsen, S.B.; Gao, S.; Ostenfeld, M.S.; Borre, M.; et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int. J. Cancer 2011, 128, 1327–1334. [Google Scholar] [CrossRef] [PubMed]
- Uesugi, A.; Kozaki, K.; Tsuruta, T.; Furuta, M.; Morita, K.; Imoto, I.; Omura, K.; Inazawa, J. The tumor suppressive microRNA miR-218 targets the mtor component rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res. 2011, 71, 5765–5778. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Zou, D.; Li, Z.; Luo, M.; Dong, L.; Wang, B.; Yin, H.; Ma, Y.; Liu, C.; Wang, F.; et al. MicroRNA-219-2-3p functions as a tumor suppressor in gastric cancer and is regulated by DNA methylation. PLoS ONE 2013, 8, e60369. [Google Scholar] [CrossRef] [PubMed]
- Png, K.J.; Yoshida, M.; Zhang, X.H.; Shu, W.; Lee, H.; Rimner, A.; Chan, T.A.; Comen, E.; Andrade, V.P.; Kim, S.W.; et al. MicroRNA-335 inhibits tumor reinitiation and is silenced through genetic and epigenetic mechanisms in human breast cancer. Genes Dev. 2011, 25, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, D.; Zhang, G.; Xiong, J.; Jie, Z.; Cheng, H.; Cao, Y.; Jiang, M.; Lin, L.; Le, Z.; et al. Methylation-associated silencing of microRNA-335 contributes tumor cell invasion and migration by interacting with rasa1 in gastric cancer. Am. J. Cancer Res. 2014, 4, 648–662. [Google Scholar] [PubMed]
- Dohi, O.; Yasui, K.; Gen, Y.; Takada, H.; Endo, M.; Tsuji, K.; Konishi, C.; Yamada, N.; Mitsuyoshi, H.; Yagi, N.; et al. Epigenetic silencing of miR-335 and its host gene mest in hepatocellular carcinoma. Int. J. Oncol. 2013, 42, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.K.; Li, Y.S.; Zhang, C.D.; Dai, D.Q. Up-regulation of crkl by microRNA-335 methylation is associated with poor prognosis in gastric cancer. Cancer Cell Int. 2017, 17, 28. [Google Scholar] [CrossRef] [PubMed]
- Grady, W.M.; Parkin, R.K.; Mitchell, P.S.; Lee, J.H.; Kim, Y.H.; Tsuchiya, K.D.; Washington, M.K.; Paraskeva, C.; Willson, J.K.; Kaz, A.M.; et al. Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene evl in colorectal cancer. Oncogene 2008, 27, 3880–3888. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.T.; Wang, J.L.; Du, W.; Hong, J.; Zhao, S.L.; Wang, Y.C.; Xiong, H.; Chen, H.M.; Fang, J.Y. MicroRNA 345, a methylation-sensitive microRNA is involved in cell proliferation and invasion in human colorectal cancer. Carcinogenesis 2011, 32, 1207–1215. [Google Scholar] [CrossRef] [PubMed]
- Nakaoka, T.; Saito, Y.; Saito, H. Aberrant DNA methylation as a biomarker and a therapeutic target of cholangiocarcinoma. Int. J. Mol. Sci. 2017, 18, 1111. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.W.; Chu, T.H.; Gong, N.R.; Chiang, W.F.; Yang, C.C.; Liu, C.J.; Wu, C.H.; Lin, S.C. miR-370 modulates insulin receptor substrate-1 expression and inhibits the tumor phenotypes of oral carcinoma. Oral Dis. 2013, 19, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gao, W.; Luo, J.; Tian, R.; Sun, H.; Zou, S. Methyl-CpG binding protein MBD2 is implicated in methylation-mediated suppression of miR-373 in hilar cholangiocarcinoma. Oncol. Rep. 2011, 25, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Chang, Y.; Li, P.; Guo, Y.; Zhang, K.; Gao, W. Androgen receptor is negatively correlated with the methylation-mediated transcriptional repression of miR-375 in human prostate cancer cells. Oncol. Rep. 2014, 31, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lin, R.; Li, J. Epigenetic silencing of microRNA-375 regulates pdk1 expression in esophageal cancer. Dig. Dis. Sci. 2011, 56, 2849–2856. [Google Scholar] [CrossRef] [PubMed]
- Mazar, J.; DeBlasio, D.; Govindarajan, S.S.; Zhang, S.; Perera, R.J. Epigenetic regulation of microRNA-375 and its role in melanoma development in humans. FEBS Lett. 2011, 585, 2467–2476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yan, D.L.; Yang, F.; Wang, D.D.; Chen, X.; Wu, J.Z.; Tang, J.H.; Xia, W.J. DNA methylation mediated silencing of microRNA-874 is a promising diagnosis and prognostic marker in breast cancer. Oncotarget 2017, 8, 45496–45505. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Choi, A.J.; Lee, B.H.; Ting, A.H. Identification and functional analysis of epigenetically silenced microRNAs in colorectal cancer cells. PLoS ONE 2011, 6, e20628. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.G.; Kim, T.O.; Bae, J.H.; Shim, J.W.; Kang, M.J.; Yang, K.; Ting, A.H.; Yi, J.M. Epigenetically regulated MIR941 and MIR1247 target gastric cancer cell growth and migration. Epigenetics 2014, 9, 1018–1030. [Google Scholar] [CrossRef] [PubMed]
- Dudziec, E.; Miah, S.; Choudhry, H.M.; Owen, H.C.; Blizard, S.; Glover, M.; Hamdy, F.C.; Catto, J.W. Hypermethylation of CpG islands and shores around specific microRNAs and miRtrons is associated with the phenotype and presence of bladder cancer. Clin. Cancer Res. 2011, 17, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, H.; Xie, Z.; Deng, W.; Wu, C.; Qin, B.; Hou, J.; Lu, M. Epigenetically regulated miR-449a enhances hepatitis b virus replication by targeting cAMP-responsive element binding protein 5 and modulating hepatocytes phenotype. Sci. Rep. 2016, 6, 25389. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.C.; Fang, W.H.; Lin, T.C.; Hou, H.A.; Chen, C.Y.; Tien, H.F.; Lin, L.I. MicroRNA let-7a-3 gene methylation is associated with karyotyping, CEBPA promoter methylation, and survival in acute myeloid leukemia. Leuk. Res. 2014, 38, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Katsaros, D.; de la Longrais, I.A.; Sochirca, O.; Yu, H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007, 67, 10117–10122. [Google Scholar] [CrossRef] [PubMed]
- Brueckner, B.; Stresemann, C.; Kuner, R.; Mund, C.; Musch, T.; Meister, M.; Sultmann, H.; Lyko, F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007, 67, 1419–1423. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.S.; Man, O.Y.; Tsang, C.M.; Tsao, S.W.; Tsang, R.K.; Chan, J.Y.; Ho, W.K.; Wei, W.I.; To, V.S. MicroRNA let-7 suppresses nasopharyngeal carcinoma cells proliferation through downregulating c-Myc expression. J. Cancer Res. Clin. Oncol. 2011, 137, 415–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geeleher, P.; Huang, S.R.; Gamazon, E.R.; Golden, A.; Seoighe, C. The regulatory effect of miRNAs is a heritable genetic trait in humans. BMC Genom. 2012, 13, 383. [Google Scholar]
- Lim, J.P.; Brunet, A. Bridging the transgenerational gap with epigenetic memory. Trends Genet. 2013, 29, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Liebers, R.; Rassoulzadegan, M.; Lyko, F. Epigenetic regulation by heritable RNA. PLoS Genet. 2014, 10, e1004296. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A landscape takes shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Kazanets, A.; Shorstova, T.; Hilmi, K.; Marques, M.; Witcher, M. Epigenetic silencing of tumor suppressor genes: Paradigms, puzzles, and potential. Biochim. Biophys. Acta 2016, 1865, 275–288. [Google Scholar] [PubMed]
- Zhang, W.; Xu, J. DNA methyltransferases and their roles in tumorigenesis. Biomark. Res. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. MicroRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar] [CrossRef] [PubMed]
- Duursma, A.M.; Kedde, M.; Schrier, M.; le Sage, C.; Agami, R. miR-148 targets human DNMT3b protein coding region. RNA 2008, 14, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Benetti, R.; Gonzalo, S.; Jaco, I.; Munoz, P.; Gonzalez, S.; Schoeftner, S.; Murchison, E.; Andl, T.; Chen, T.; Klatt, P.; et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 2008, 15, 998. [Google Scholar] [CrossRef] [PubMed]
- Vire, E.; Brenner, C.; Deplus, R.; Blanchon, L.; Fraga, M.; Didelot, C.; Morey, L.; Van Eynde, A.; Bernard, D.; Vanderwinden, J.M.; et al. The polycomb group protein EZH2 directly controls DNA methylation. Nature 2006, 439, 871–874. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Zhang, Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 2004, 15, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Lindahl-Allen, M.; Polytarchou, C.; Hirsch, H.A.; Tsichlis, P.N.; Struhl, K. Loss of miR-200 inhibition of SUZ12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 2010, 39, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Peruzzi, P.; Bronisz, A.; Nowicki, M.O.; Wang, Y.; Ogawa, D.; Price, R.; Nakano, I.; Kwon, C.H.; Hayes, J.; Lawler, S.E.; et al. MicroRNA-128 coordinately targets polycomb repressor complexes in glioma stem cells. Neuro Oncol. 2013, 15, 1212–1224. [Google Scholar] [CrossRef] [PubMed]
- Kleer, C.G.; Cao, Q.; Varambally, S.; Shen, R.; Ota, I.; Tomlins, S.A.; Ghosh, D.; Sewalt, R.G.; Otte, A.P.; Hayes, D.F.; et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11606–11611. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, I.M.; Halvorsen, O.J.; Collett, K.; Stefansson, I.M.; Straume, O.; Haukaas, S.A.; Salvesen, H.B.; Otte, A.P.; Akslen, L.A. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006, 24, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Sander, S.; Bullinger, L.; Klapproth, K.; Fiedler, K.; Kestler, H.A.; Barth, T.F.; Moller, P.; Stilgenbauer, S.; Pollack, J.R.; Wirth, T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 2008, 112, 4202–4212. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.M.; Liang, G.; Liu, C.C.; Wolff, E.M.; Tsai, Y.C.; Ye, W.; Zhou, X.; Jones, P.A. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009, 69, 2623–2629. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Cao, Q.; Mani, R.S.; Shankar, S.; Wang, X.; Ateeq, B.; Laxman, B.; Cao, X.; Jing, X.; Ramnarayanan, K.; et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008, 322, 1695–1699. [Google Scholar] [CrossRef] [PubMed]
- Di Croce, L.; Helin, K. Transcriptional regulation by polycomb group proteins. Nat. Struct. Mol. Biol. 2013, 20, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Li, J.; Song, L. Bmi-1, stem cells and cancer. Acta Biochim. Biophys. Sin. (Shanghai) 2009, 41, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Godlewski, J.; Nowicki, M.O.; Bronisz, A.; Williams, S.; Otsuki, A.; Nuovo, G.; Raychaudhury, A.; Newton, H.B.; Chiocca, E.A.; Lawler, S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008, 68, 9125–9130. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, R.; Nicoloso, M.; Arvizo, R.; Wang, E.; Cortez, A.; Rossi, S.; Calin, G.A.; Mukherjee, P. miR-15a and miR-16 control Bmi-1 expression in ovarian cancer. Cancer Res. 2009, 69, 9090–9095. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Kaneuchi, M.; Watari, H.; Hamada, J.; Sudo, S.; Ju, J.; Sakuragi, N. MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol. Cancer 2011, 10, 10–1186. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.; Gao, X.; Li, G.; Fu, H.; Cui, D.; Liu, H.; Jin, W.; Zhang, Y. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res. 2013, 73, 6046–6055. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Q.; Niu, W.Y.; Li, Y.P.; Huang, H.B.; Zhan, R. miR-203 inhibits cell growth and regulates G1/S transition by targeting Bmi-1 in myeloma cells. Mol. Med. Rep. 2016, 14, 4795–4801. [Google Scholar] [CrossRef] [PubMed]
- Van der Vlag, J.; Otte, A.P. Transcriptional repression mediated by the human polycomb-group protein eed involves histone deacetylation. Nat. Genet. 1999, 23, 474–478. [Google Scholar] [CrossRef] [PubMed]
- Witt, O.; Deubzer, H.E.; Milde, T.; Oehme, I. Hdac family: What are the cancer relevant targets? Cancer Lett. 2009, 277, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Noonan, E.J.; Place, R.F.; Pookot, D.; Basak, S.; Whitson, J.M.; Hirata, H.; Giardina, C.; Dahiya, R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 2009, 28, 1714–1724. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.H.; Chang, Y.G.; Kim, M.G.; Jung, K.H.; Kim, J.K.; Bae, H.J.; Eun, J.W.; Shen, Q.; Kim, S.J.; Kwon, S.H.; et al. miR-145 functions as a tumor suppressor by directly targeting histone deacetylase 2 in liver cancer. Cancer Lett. 2013, 335, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, S.K.; Volinia, S.; Costinean, S.; Galasso, M.; Neinast, R.; Santhanam, R.; Parthun, M.R.; Perrotti, D.; Marcucci, G.; Garzon, R.; et al. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Eμ-miR-155 transgenic mouse model. Proc. Natl. Acad. Sci. USA 2012, 109, 20047–20052. [Google Scholar] [CrossRef] [PubMed]
- Canzio, D.; Larson, A.; Narlikar, G.J. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol. 2014, 24, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Dialynas, G.K.; Vitalini, M.W.; Wallrath, L.L. Linking heterochromatin protein 1 (HP1) to cancer progression. Mutat. Res. 2008, 647, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Huang, F.; Zhang, D.; Ju, J.; Wu, X.B.; Wang, Y.; Wu, Y.; Nie, M.; Li, Z.; Ma, C.; et al. Heterochromatin protein HP1γ promotes colorectal cancer progression and is regulated by miR-30a. Cancer Res. 2015, 75, 4593–4604. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yan, T.; Liu, Z.; Wang, J.; Lu, Y.; Li, D.; Liang, W. MicroRNA-137 is negatively associated with clinical outcome and regulates tumor development through EZH2 in cervical cancer. J. Cell Biochem. 2018, 119, 938–947. [Google Scholar] [CrossRef] [PubMed]
- Takata, A.; Otsuka, M.; Yoshikawa, T.; Kishikawa, T.; Hikiba, Y.; Obi, S.; Goto, T.; Kang, Y.J.; Maeda, S.; Yoshida, H.; et al. MicroRNA-140 acts as a liver tumor suppressor by controlling NF-κB activity by directly targeting DNA methyltransferase 1 (DNMT1) expression. Hepatology 2013, 57, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Wang, Y.; Xi, Y.; Kudo, K.; Bruheim, S.; Botchkina, G.I.; Gavin, E.; Wan, Y.; Formentini, A.; Kornmann, M.; et al. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene 2009, 28, 4065–4074. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.K.; Tsang, W.P.; Ng, S.S.; Jin, H.C.; Yu, J.; Li, J.J.; Rocken, C.; Ebert, M.P.; Kwok, T.T.; Sung, J.J. MicroRNA-143 targets DNA methyltransferases 3a in colorectal cancer. Br. J. Cancer 2009, 101, 699–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsui, M.; Chu, Y.; Zhang, H.; Gagnon, K.T.; Shaikh, S.; Kuchimanchi, S.; Manoharan, M.; Corey, D.R.; Janowski, B.A. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013, 41, 10086–10109. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Xia, J.; Zuo, J.; Jin, S.; Zhou, H.; Yao, L.; Huang, H.; Han, Z. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in gastric cancer. Med. Oncol. 2012, 29, 2701–2709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tang, H.; Wang, Z.; Zhang, B.; Liu, W.; Lu, H.; Xiao, L.; Liu, X.; Wang, R.; Li, X.; et al. miR-185 targets the DNA methyltransferases 1 and regulates global DNA methylation in human glioma. Mol. Cancer 2011, 10, 124. [Google Scholar] [CrossRef] [PubMed]
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.J.; Jung, K.H.; Eun, J.W.; Shen, Q.; Kim, H.S.; Park, S.J.; Shin, W.C.; Yang, H.D.; Park, W.S.; Lee, J.Y.; et al. MicroRNA-221 governs tumor suppressor hdac6 to potentiate malignant progression of liver cancer. J. Hepatol. 2015, 63, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.W.; Wentzel, E.A.; Mendell, J.T. A hexanucleotide element directs microRNA nuclear import. Science 2007, 315, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Villeneuve, L.M.; Morris, K.V.; Rossi, J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006, 13, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Janowski, B.A.; Huffman, K.E.; Schwartz, J.C.; Ram, R.; Nordsell, R.; Shames, D.S.; Minna, J.D.; Corey, D.R. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat. Struct. Mol. Biol. 2006, 13, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Huang, V.; Zheng, J.; Qi, Z.; Wang, J.; Place, R.F.; Yu, J.; Li, H.; Li, L.C. Ago1 interacts with RNA polymerase ii and binds to the promoters of actively transcribed genes in human cancer cells. PLoS Genet. 2013, 9, e1003821. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhang, B.; Wu, T.; Skogerbo, G.; Zhu, X.; Guo, X.; He, S.; Chen, R. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol. Biol. 2009, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Majid, S.; Dar, A.A.; Saini, S.; Yamamura, S.; Hirata, H.; Tanaka, Y.; Deng, G.; Dahiya, R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer 2010, 116, 5637–5649. [Google Scholar] [CrossRef] [PubMed]
- Zardo, G.; Ciolfi, A.; Vian, L.; Starnes, L.M.; Billi, M.; Racanicchi, S.; Maresca, C.; Fazi, F.; Travaglini, L.; Noguera, N.; et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood 2012, 119, 4034–4046. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Saetrom, P.; Snove, O., Jr.; Rossi, J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16230–16235. [Google Scholar] [CrossRef] [PubMed]
- Place, R.F.; Li, L.C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613. [Google Scholar] [CrossRef] [PubMed]
- Younger, S.T.; Corey, D.R. Transcriptional gene silencing in mammalian cells by miRna mimics that target gene promoters. Nucleic Acids Res. 2011, 39, 5682–5691. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Roth, A.; Yu, M.; Morris, R.; Bersani, F.; Rivera, M.N.; Lu, J.; Shioda, T.; Vasudevan, S.; Ramaswamy, S.; et al. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev. 2013, 27, 2543–2548. [Google Scholar] [CrossRef] [PubMed]
- Huang, V.; Place, R.F.; Portnoy, V.; Wang, J.; Qi, Z.; Jia, Z.; Yu, A.; Shuman, M.; Yu, J.; Li, L.C. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012, 40, 1695–1707. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.V.; Chan, S.W.; Jacobsen, S.E.; Looney, D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 2004, 305, 1289–1292. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, M.S.; Villeneuve, L.M.; Ehsani, A.; Amarzguioui, M.; Aagaard, L.; Chen, Z.X.; Riggs, A.D.; Rossi, J.J.; Morris, K.V. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 2006, 12, 256–262. [Google Scholar] [CrossRef] [PubMed]
- White, R.J. Rna polymerase III transcription and cancer. Oncogene 2004, 23, 3208–3216. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.K.; Yochmowitz, M.G.; Knight, W.A., 3rd; McGuire, W.L. The value of estrogen and progesterone receptors in the treatment of breast cancer. Cancer 1980, 46, 2884–2888. [Google Scholar] [CrossRef]
- Starnes, L.M.; Sorrentino, A.; Pelosi, E.; Ballarino, M.; Morsilli, O.; Biffoni, M.; Santoro, S.; Felli, N.; Castelli, G.; De Marchis, M.L.; et al. NFI-A directs the fate of hematopoietic progenitors to the erythroid or granulocytic lineage and controls β-globin and G-CSF receptor expression. Blood 2009, 114, 1753–1763. [Google Scholar] [CrossRef] [PubMed]
- Janowski, B.A.; Younger, S.T.; Hardy, D.B.; Ram, R.; Huffman, K.E.; Corey, D.R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007, 3, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Bjornsson, H.T.; Brown, L.J.; Fallin, M.D.; Rongione, M.A.; Bibikova, M.; Wickham, E.; Fan, J.B.; Feinberg, A.P. Epigenetic specificity of loss of imprinting of the IGF2 gene in wilms tumors. J. Natl. Cancer Inst. 2007, 99, 1270–1273. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Qin, Z.; Hu, Z.; Guan, X.; Wang, Y.; He, Y.; Xue, J.; Liu, X.; Chen, J.; Dai, J.; et al. Genetic variation in a hsa-let-7 binding site in RAD52 is associated with breast cancer susceptibility. Carcinogenesis 2013, 34, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Pandolfi, P.P. The epitranscriptome of noncoding RNAs in cancer. Cancer Discov. 2017, 7, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.; Zander, S.; Gutschner, T. The dark side of the epitranscriptome: Chemical modifications in long non-coding RNAs. Int. J. Mol. Sci. 2017, 18, 2387. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, X.; Yu, S.; Jeong, K.J.; Zhou, Z.; Han, L.; Tsang, Y.H.; Li, J.; Chen, H.; Mangala, L.S.; et al. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res. 2017, 27, 1112–1125. [Google Scholar] [CrossRef] [PubMed]
- Monnier, P.; Martinet, C.; Pontis, J.; Stancheva, I.; Ait-Si-Ali, S.; Dandolo, L. H19 lncRNA controls gene expression of the imprinted gene network by recruiting MBD1. Proc. Natl. Acad. Sci. USA 2013, 110, 20693–20698. [Google Scholar] [CrossRef] [PubMed]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Chiyomaru, T.; Fukuhara, S.; Saini, S.; Majid, S.; Deng, G.; Shahryari, V.; Chang, I.; Tanaka, Y.; Enokida, H.; Nakagawa, M.; et al. Long non-coding rna hotair is targeted and regulated by miR-141 in human cancer cells. J. Biol. Chem. 2014, 289, 12550–12565. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Yao, J.; An, Y.; Chen, X.; Chen, W.; Wu, D.; Luo, B.; Yang, Y.; Jiang, Y.; Sun, D.; et al. LncRNA HOTAIR acts a competing endogenous RNA to control the expression of Notch3 via sponging miR-613 in pancreatic cancer. Oncotarget 2017, 8, 32905–32917. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]


| miRNA | Cancer Type | Epigenetic Modification | Target | Reference |
|---|---|---|---|---|
| miR-1 | Hepatocellular, liver, colorectal, lung | DMhyper | FOXP1, MET, HDAC4, Pim1 | [74,121,122] |
| miR-9 | Breast, ovarian, pancreatic, multiple myeloma, renal, gastric, hepatocellular, colorectal, melanoma, head and neck, multiple myeloma, lung | DMhyper | CCNG1, IL-6, AP3B1, TC10, ONECUT2, IGF2BP1, MYO1D, ANXA2 | [44,47,58,67,75,123,124,125] |
| miR-10a | Gastric, bladder, hepatocellular | DMhyper | HOXA1 | [69,126,127] |
| miR-10b | Gastric, hepatocellular | DMhyper | [70,127] | |
| miR-15a/16 | Chronic lymphocytic leukemia, mantle cell lymphoma | HDA | BCL2, MCL1 | [109,110] |
| miR-17-92 | Colorectal | HDA | PTEN, BCL2L11, CDKN1A | [128] |
| miR-21 | Ovarian, prostate, colorectal | DMhypo, DMhyper, HMT | ITGB4 | [129,130,131] |
| miR-23a-27a | Hepatocellular | DMhypo | [78] | |
| miR-24 | Nasopharyngeal | DMhyper | [132] | |
| miR-29a/b | B-cell Lymphoma, chronic lymphocytic leukemia, acute myeloid leukemia, lung | HMT, HDA | MCL1, DNMT3A-B | [31,32,109,133,134] |
| miR-31 | Melanoma, prostate, breast | HMT, DMhyper, HDA | SRC, RAB27A, MAP3K14, MET, E2F1, E2F2, EXO1, FOXM1, MCM2, E2F6, BMI-1 | [120,135,136,137,138,139] |
| miR-33b | Gastric | DMhyper | [72] | |
| miR-34a | Lung, breast, colon, kidney, bladder, pancreatic cancer cells, melanoma | DMhyper | CDK6 | [76,140,141] |
| miR-34b/c | Gastric, ovarian, lung, colon, melanoma, head and neck, breast, non-small cell lung, neuroblastoma, hepatocellular, pleural mesothelioma, oral | DMhyper | MYC, CDK6, E2F3 | [45,68,76,141,142,143,144,145] |
| miR-101 | Hepatocellular | HMT | [118] | |
| miR-106b-25-93 | Hepatocellular | DMhypo | [78] | |
| miR-107 | Pancreatic | DMhyper | CDK6 | [146] |
| miR-124 | Colon, gastric, hematological, cervical, glioblastoma cells, breast, prostate, neuroblastoma, pancreatic, colorectal, non-small cell lung, acute lymphoblastic leukemia, hepatocellular, renal | DMhyper | BCL2, CDK6, VIM, SMYD3, IQGAP1, RAC1 | [45,59,73,77,142,144,147,148,149,150,151,152] |
| miR-125b | Breast, hepatocellular | HMT, DMhyper | PGF | [45,118,153] |
| miR-126 | Bladder, malignant pleural mesothelioma, colorectal, non-small cell lung | DMhyper, HDA | VEGF | [154,155,156,157] |
| miR-127 | Prostate, bladder, colon, breast, clear cell renal cell carcinoma | DMhyper, HDA | DAPK1, BCL6 | [42,45,158] |
| miR-129 | Gastric, endometrial, colorectal, hepatocellular, hematological | DMhyper | SOX4 | [68,159,160,161,162,163] |
| miR-132 | Pancreas, prostate, breast | DMhyper, HDA | TALIN2, HB-EGF | [45,164,165] |
| miR-133b | Colorectal | DMhyper | [166] | |
| miR-137 | Head and neck squamous cells, colorectal, glioblastoma cells, prostate, multiple myeloma, gastric, oral, hepatocellular cells | DMhyper | CDK6, E2F6, LSD-1, ASCT2, AURKA | [98,145,151,167,168,169,170,171] |
| miR-139 | Hepatocellular, non-small cell lung | HMT | ROCK2 | [172,173] |
| miR-141 | Clear cell renal cell carcinoma | DMhyper, HDA | TET1, TET3, ZEB1 | [158,174] |
| miR-143 | Leukemia | DMhyper | MLL-AF4 | [175] |
| miR-145 | Prostate, lung adenocarcinoma, non-small cell carcinoma, clear cell renal cell carcinoma | DMhyper, HDA | TNFSF10, MUCIN1 | [158,176,177,178,179] |
| miR-148a | Colorectal, melanoma, head and neck, breast, pancreas, hepatocellular | DMhyper | TGIF2 | [44,78] |
| miR-149 | Breast | DMhyper | NDST1 | [57] |
| miR-152 | Endometrium, bladder cancer cells, prostate, breast cancer cells | DMhyper | DNMT1, E2F3, MET, RICTOR | [34,126,171,180,181] |
| miR-155 | Breast, prostate | HDA, DMhyper | [113,171] | |
| miR-181a/b | Prostate | HMT, DMhyper, HDA | RING2 | [119] |
| miR-181c | Gastric, prostate, glioblastoma cells | DMhyper | NOTCH4, KRAS, NOTCH2 | [65,182] |
| miR-191 | Breast, hepatocellular | DMhypo | TIMP3 | [45,183] |
| miR-192 | Pancreatic ductal adenocarcinoma | DMhyper | SERPINE1 | [61] |
| miR-193a | Hepatocellular, acute myeloid leukemia, bladder, breast, oral | DMhyper, DMhypo | E2F6, SRSF2, PLAU, HIC2 | [45,145,184,185,186] |
| miR-193b | Prostate | DMhyper, HDA | [187,188] | |
| miR-195/497 | Hepatocellular | DMhyper | [78] | |
| miR-196b | Gastric, prostate, hepatocellular | DMhyper, DMhypo | [127,131,189] | |
| miR-199a | Testicular, ovarian | DMhyper | PODXL, DDR1 | [190,191] |
| miR-200a/b/429 | Hepatocellular, prostate, gastric, glioblastoma, pancreatic, bladder | HMT, DMhyper, HDA, DMhypo | BMI1, RING2 | [63,71,118,119,126,192] |
| miR-200c/141 | Colon, breast, lung, prostate, non-small cell lung | HMT, DMhyper, HDA | DNMT3A TET1, TET3, BMI1, RING2, SOX2, ZEB1, DNMT3A | [119,174,193,194,195] |
| miR-203 | Hematological, hepatocellular, endometrial, ovarian, prostate, oral | DMhyper, DMhypo, HMT, HDA | ABCE1, BMI1, SOX4 | [77,119,129,145,196,197] |
| miR-205 | Bladder, prostate, ovarian | DMhypo, DMhyper | BCL2L2 | [129,131,139,198] |
| miR-218 | Oral squamous cell carcinoma | DMhyper | RICTOR | [199] |
| miR-219a | Gastric, endometrial | DMhyper | [196,200] | |
| miR-221 | Hepatocellular | DMhypo | MDM2 | [81] |
| miR-224 | Hepatocellular | HDA, HAT | [116] | |
| miR-335 | Breast, hepatocellular, gastric | DMhyper | RASA1, CRKL | [201,202,203,204] |
| miR-342 | Colorectal | DMhyper | [205] | |
| miR-345 | Colorectal | DMhyper | BAG3 | [206] |
| miR-370 | Cholangiocarcinoma, oral squamous cells | DMhyper | IRS1 | [207,208] |
| miR-373 | Cholangiocarcinoma | DMhyper, HDA | [209] | |
| miR-375 | Esophagus, melanoma, prostate, hepatocellular, breast | DMhyper | RASFF1(A), PDK1 | [45,78,210,211,212] |
| miR-376c | Cholangiocarcinoma | DMhyper | [207] | |
| miR-378 | Hepatocellular | DMhyper | [78] | |
| miR-449a/b | Osteosarcoma cell line, breast cell line, hepatocellular | HMT, HDA | CDK6, CDC25A, C-MET | [115,117] |
| miR-512 | Gastric | DMhyper, HDA | [42] | |
| miR-514 | Clear cell renal cell carcinoma | DMhyper, HDA | [158] | |
| miR-585 | Oral squamous cell carcinoma | DMhyper | [199] | |
| miR-596 | Endometrial | DMhyper | [196] | |
| miR-615 | Pancreatic ductal adenocarcinoma | DMhypo | IGF2 | [60,131] |
| miR-618 | Endometrial | DMhyper | [196] | |
| miR-874 | Breast | DMhyper | [213] | |
| miR-941 | Colorectal cells | DMhyper | [214,215] | |
| miR-1224 | Bladder | DMhyper | [214,216] | |
| miR-1237 | Colorectal cells | DMhyper | [214] | |
| miR-1247 | Colorectal and gastric cells, pancreatic, non-small cell lung | DMhyper | RARA, STX1B, RCC2 | [62,214,215,217] |
| Let-7a | Ovarian, acute myeloid leukemia, lung, nasopharyngeal carcinoma cells | DMhyper, DMhypo | C-MYC | [218,219,220,221] |
| Let-7c | Hepatocellular | HMT | [118] |
| MicroRNAs | Target | Cancer Type | Reference |
|---|---|---|---|
| miR-15a/16-1 | BMI | Ovarian | [244] |
| miR-26a | EZH2 | Burkit lymphoma | [238] |
| miR-29a/b | DNMT3A-B, DNMT1 | Lung, acute myeloid leukemia, hepatocellular | [31,32,228] |
| miR-30a | HP1γ | Colorectal | [255] |
| miR-101 | EZH2 | Prostate, bladder transitional cell carcinoma | [239,240] |
| miR-128 | BMI, SUZ12 | Glioma | [235,243] |
| miR-137 | EZH2 | Cervical | [256] |
| miR-140 | DNMT1, HDAC4 | Hepatocellular, osteosarcoma, colorectal | [257,258] |
| miR-143 | DNMT3A | Colorectal | [259] |
| miR-145 | HDAC2 | Hepatocellular | [260] |
| miR-148 | DNMT3B | Cervical cancer cells | [229] |
| miR-148a | DNMT1 | Cholangiocarcinoma, gastric | [34,261] |
| miR-152 | DNMT1 | Cholangioarcinoma, breast | [34,181] |
| miR-155 | HDAC4 | B-cells lymphoma | [252] |
| miR-185 | DNMT1 | Glioma | [262] |
| miR-194 | BMI | Endometrial | [245] |
| miR-200b | SUZ12, BMI | Breast, hepatocellular | [192,234] |
| miR-200c | BMI | Breast | [263] |
| miR-203 | BMI | Multiple myeloma | [247] |
| miR-218 | BMI | Glioma | [246] |
| miR-221 | HDAC6 | Liver | [264] |
| miR-449a | HDAC1 | Prostate | [250] |
| miR-K12-4-5p | RBL2 | Kaposi’s sarcoma-associated herpesvirus | [33] |
| MicroRNA | Target | TGS/TGA | Reference |
|---|---|---|---|
| miR-10a | HOXD4 | TGS | [269] |
| miR-205 | IL24 | TGA | [270] |
| miR-205 | IL32 | TGA | [270] |
| miR-223 | NFI-A | TGS | [271] |
| miR-320 | POLR3D | TGS | [272] |
| miR-373 | CDH1 | TGA | [273] |
| miR-373 | CSDC2 | TGA | [273] |
| miR-423 (synthetic) | PR | TGS | [274] |
| miR-483 | IGF2 | TGA | [275] |
| miR-589 | COX2 | TGA | [260] |
| miR-774 | Cnnb1 | TGA | [276] |
| miR-1186 | Cnnb1 | TGA | [276] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramassone, A.; Pagotto, S.; Veronese, A.; Visone, R. Epigenetics and MicroRNAs in Cancer. Int. J. Mol. Sci. 2018, 19, 459. https://doi.org/10.3390/ijms19020459
Ramassone A, Pagotto S, Veronese A, Visone R. Epigenetics and MicroRNAs in Cancer. International Journal of Molecular Sciences. 2018; 19(2):459. https://doi.org/10.3390/ijms19020459
Chicago/Turabian StyleRamassone, Alice, Sara Pagotto, Angelo Veronese, and Rosa Visone. 2018. "Epigenetics and MicroRNAs in Cancer" International Journal of Molecular Sciences 19, no. 2: 459. https://doi.org/10.3390/ijms19020459
APA StyleRamassone, A., Pagotto, S., Veronese, A., & Visone, R. (2018). Epigenetics and MicroRNAs in Cancer. International Journal of Molecular Sciences, 19(2), 459. https://doi.org/10.3390/ijms19020459