Organ and Tissue-Specific Localisation of Selected Cell Wall Epitopes in the Zygotic Embryo of Brachypodium distachyon
Abstract
:1. Introduction
2. Results and Discussion
2.1. The Morphological and Histological Features of Brachypodium Embryos
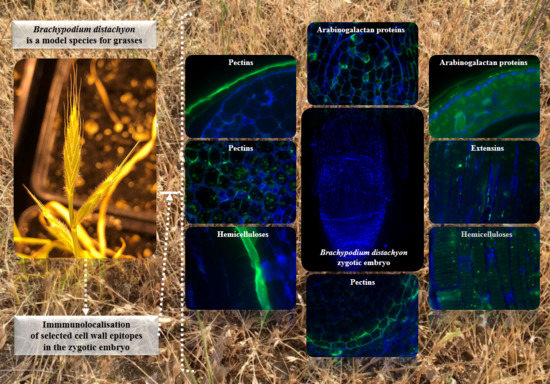
2.2. AGP, Extensin, Pectin, and Hemicellulose Epitopes in Various Tissues and Organs of Brachypodium Embryos
3. Materials and Methods
3.1. Plant Material and Histological Procedures
3.2. Immunocytochemistry
3.3. TEM
4. Conclusions
- Among analysed AGP epitopes JIM13, JIM8, and LM2 were localised in cell walls of the embryo surface tissues, as well as some of them in the seed coat; and MAC207 and JIM16 were detected in cytoplasmic compartments.
- Extensins are localised in the outer periclinal walls of embryo surface tissues. Our results suggest that in Brachypodium, the antibody LM13 can be used as a negative marker of epidermal cells of the embryonic leaf and that the LM25 antibody can be used as the positive marker of embryonic root epidermis cells, as well as the root cap in Brachypodium. These results demonstrate that the distribution of analysed cell wall epitopes in mature zygotic embryos of Brachypodium is tissue- and organ specific.
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AGP | Arabinogalactan protein |
| BSA | Bovine serum albumin |
| CRISPR | Clustered regularly-interspaced short palindromic repeats |
| FCS | Foetal calf serum |
| GA | Glutaraldehyde |
| PBS | Phosphate-buffered saline |
| PFA | Paraformaldehyde |
| RT | Room temperature |
| TEM | Transmission electron microscope |
References
- Showalter, A.M. Structure and function of plant cell wall proteins. Plant Cell 1993, 5, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Villarreal, M.; Aispuro-Hernández, E.; Vargas-Arispuro, I.; Ángel Martínez-Téllez, M. Plant Cell Wall Polymers: Function, Structure and Biological Activity of Their Derivatives. In Materials Science; Gomes, A.D.S., Ed.; INTECH: Rijeka, Croatia, 2012. [Google Scholar]
- Stafford, H.A. Plant Cell Wall Polymers: Biogenesis and Biodegradation; Lewis, N.G., Paice, M.G., Eds.; American Chemical Society: Washington, DC, USA, 1989; p. xii. 676p. [Google Scholar]
- Schadel, C. Cell-Wall Hemicelluloses as Mobile Carbon Stores in Plants. Ph.D. Thesis, Universität Basel, Basel, Switzerland, 2009. [Google Scholar]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Evert, R.F. Esau’s Plant Anatomy: Meristems, Cells, and Tissues of the Plant Body: Their Structure, Function, and Development; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006. [Google Scholar]
- Labavitch, J.M.; Freeman, L.E.; Albersheim, P. Structure of plant cell walls. Purification and characterization of a beta-1,4-galactanase which degrades a structural component of the primary cell walls of dicots. J. Biol. Chem. 1976, 251, 5904–5910. [Google Scholar] [PubMed]
- Draper, J.; Mur, L.A.; Jenkins, G.; Ghosh-Biswas, G.C.; Bablak, P.; Hasterok, R.; Routledge, A.P. Brachypodium distachyon a new model system for functional genomics in grasses. Plant Physiol. 2001, 127, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Francin-Allami, M.; Merah, K.; Albenne, C.; Rogniaux, H.; Pavlovic, M.; Lollier, V.; Sibout, R.; Guillon, F.; Jamet, E.; Larre, C. Cell wall proteomic of Brachypodium distachyon grains: A focus on cell wall remodeling proteins. Proteomics 2015, 15, 2296–2306. [Google Scholar] [CrossRef] [PubMed]
- Christensen, U.; Alonso-Simon, A.; Scheller, H.V.; Willats, W.G.; Harholt, J. Characterization of the primary cell walls of seedlings of Brachypodium distachyon-a potential model plant for temperate grasses. Phytochemistry 2010, 71, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Salazar, A.; Zabotina, O.A.; Hong, M. Structure and dynamics of Brachypodium primary cell wall polysaccharides from two-dimensional 13C solid-state nuclear magnetic resonance spectroscopy. Biochemistry 2014, 53, 2840–2854. [Google Scholar] [CrossRef] [PubMed]
- Marriott, P.E.; Sibout, R.; Lapierre, C.; Fangel, J.U.; Willats, W.G.; Hofte, H.; Gomez, L.D.; McQueen-Mason, S.J. Range of cell-wall alterations enhance saccharification in Brachypodium distachyon mutants. Proc. Natl. Acad. Sci. USA 2014, 111, 14601–14606. [Google Scholar] [CrossRef] [PubMed]
- Wolny, E.; Braszewska-Zalewska, A.; Hasterok, R. Spatial distribution of epigenetic modifications in Brachypodium distachyon embryos during seed maturation and germination. PLoS ONE 2014, 9, e101246. [Google Scholar] [CrossRef] [PubMed]
- Tillich, H. Vergleichend morphologische Untersuchungen zur Identitat der Gramineen-Primarwurzel. Flora 1977, 166, 415–421. [Google Scholar] [CrossRef]
- Dolan, L.; Janmaat, K.; Willemsen, V.; Linstead, P.; Poethig, S.; Roberts, K.; Scheres, B. Cellular organisation of the Arabidopsis thaliana root. Development 1993, 119, 71–84. [Google Scholar] [PubMed]
- Verdeil, J.L.; Alemanno, L.; Niemenak, N.; Tranbarger, T.J. Pluripotent versus totipotent plant stem cells: Dependence versus autonomy? Trends Plant Sci. 2007, 12, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Jouannic, S.; Lartaud, M.; Herve, J.; Collin, M.; Orieux, Y.; Verdeil, J.L.; Tregear, J.W. The shoot apical meristem of oil palm (Elaeis guineensis; Arecaceae): Developmental progression and dynamics. Ann. Bot. 2011, 108, 1477–1487. [Google Scholar] [CrossRef] [PubMed]
- Marín-Méndez, W.; Sanchéz-Chacón, E.; Gatica-Arias, A.M.; Ramírez Fonseca, P.; Freer-Bustamante, E.; Valdez-Melara, M. Ultrastructure and histology of organogenesis induced from shoot tips of maize (Zea mays, Poaceae). Rev. Biol. Trop. 2009, 57, 129–139. [Google Scholar]
- Bennici, A.; Tani, C. Ultrastructural effects of salinity in Nicotiana bigelovii var. bigelovii callus and Allium cepa roots. Caryologia 2009, 62, 124–133. [Google Scholar]
- Rumyantseva, N.I. Arabinogalactan proteins: Involvement in plant growth and morphogenesis. Biochemistry 2005, 70, 1073–1085. [Google Scholar] [CrossRef] [PubMed]
- Joosen, R.V.; Arends, D.; Willems, L.A.; Ligterink, W.; Jansen, R.C.; Hilhorst, H.W. Visualizing the genetic landscape of Arabidopsis seed performance. Plant Physiol. 2012, 158, 570–589. [Google Scholar] [CrossRef] [PubMed]
- Joosen, R.V.L.; Ligterink, W.; Dekkers, B.J.W.; Hilhorst, H.W.M. Visualization of molecular processes associated with seed dormancy and germination using MapMan. Seed Sci. Res. 2011, 21, 143–152. [Google Scholar] [CrossRef]
- Cannesan, M.A.; Durand, C.; Burel, C.; Gangneux, C.; Lerouge, P.; Ishii, T.; Laval, K.; Follet-Gueye, M.L.; Driouich, A.; Vicre-Gibouin, M. Effect of arabinogalactan proteins from the root caps of pea and Brassica napus on Aphanomyces euteiches zoospore chemotaxis and germination. Plant Physiol. 2012, 159, 1658–1670. [Google Scholar] [CrossRef] [PubMed]
- Van Hengel, A.J.; Roberts, K. AtAGP30, an arabinogalactan-protein in the cell walls of the primary root, plays a role in root regeneration and seed germination. Plant J. 2003, 36, 256–270. [Google Scholar] [CrossRef] [PubMed]
- Brady, J.D.; Sadler, I.H.; Fry, S.C. Pulcherosine, an oxidatively coupled trimer of tyrosine in plant cell walls: Its role in cross-link formation. Phytochemistry 1998, 47, 349–353. [Google Scholar] [CrossRef]
- Cannon, M.C.; Terneus, K.; Hall, Q.; Tan, L.; Wang, Y.; Wegenhart, B.L.; Chen, L.; Lamport, D.T.; Chen, Y.; Kieliszewski, M.J. Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 2008, 105, 2226–2231. [Google Scholar] [CrossRef] [PubMed]
- Lamport, D.T.; Kieliszewski, M.J.; Chen, Y.; Cannon, M.C. Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 2011, 156, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Takac, T.; Burbach, C.; Menzel, D.; Samaj, J. Developmental localization and the role of hydroxyproline rich glycoproteins during somatic embryogenesis of banana (Musa spp. AAA). BMC Plant Biol. 2011, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Sujkowska-Rybkowska, M.; Borucki, W. Accumulation and localization of extensin protein in apoplast of pea root nodule under aluminum stress. Micron 2014, 67, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Fan, W.; Li, X.; Chen, H.; Takac, T.; Samajova, O.; Fabrice, M.R.; Xie, L.; Ma, J.; Samaj, J.; et al. Expression and distribution of extensins and AGPs in susceptible and resistant banana cultivars in response to wounding and Fusarium oxysporum. Sci. Rep. 2017, 7, 42400. [Google Scholar] [CrossRef] [PubMed]
- Szatanik-Kloc, A.; Szerement, J.; Jozefaciuk, G. The role of cell walls and pectins in cation exchange and surface area of plant roots. J. Plant Physiol. 2017, 215, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Prade, R.A.; Zhan, D.; Ayoubi, P.; Mort, A.J. Pectins, pectinases and plant-microbe interactions. Biotechnol. Genet. Eng. Rev. 1999, 16, 361–391. [Google Scholar] [CrossRef] [PubMed]
- Carpita, N.C.; Defernez, M.; Findlay, K.; Wells, B.; Shoue, D.A.; Catchpole, G.; Wilson, R.H.; McCann, M.C. Cell wall architecture of the elongating maize coleoptile. Plant Physiol. 2001, 127, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Verhertbruggen, Y.; Marcus, S.E.; Chen, J.; Knox, J.P. Cell wall pectic arabinans influence the mechanical properties of Arabidopsis thaliana inflorescence stems and their response to mechanical stress. Plant Cell Physiol. 2013, 54, 1278–1288. [Google Scholar] [CrossRef] [PubMed]
- Verhertbruggen, Y.; Marcus, S.E.; Haeger, A.; Verhoef, R.; Schols, H.A.; McCleary, B.V.; McKee, L.; Gilbert, H.J.; Knox, J.P. Developmental complexity of arabinan polysaccharides and their processing in plant cell walls. Plant J. 2009, 59, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Gomez, L.D.; Steele-King, C.G.; Jones, L.; Foster, J.M.; Vuttipongchaikij, S.; McQueen-Mason, S.J. Arabinan metabolism during seed development and germination in Arabidopsis. Mol. Plant 2009, 2, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Gibeaut, D.M.; Pauly, M.; Bacic, A.; Fincher, G.B. Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 2005, 221, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Dourado, F.; Barros, A.; Mota, M.; Coimbra, M.A.; Gama, F.M. Anatomy and cell wall polysaccharides of almond (Prunus dulcis D. A. Webb) seeds. J. Agric. Food Chem. 2004, 52, 1364–1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, I.; Andersson, R.; Westerlund, E.; Andersson, R.; Aman, P. Structural features of an arabinan fragment isolated from the water-soluble fraction of dehulled rapeseed. Carbohydr. Res. 1996, 281, 161–172. [Google Scholar] [CrossRef]
- Navarro, D.A.; Cerezo, A.S.; Stortz, C.A. NMR spectroscopy and chemical studies of an arabinan-rich system from the endosperm of the seed of Gleditsia triacanthos. Carbohydr. Res. 2002, 337, 255–263. [Google Scholar] [CrossRef]
- Wang, T.; Hong, M. Solid-state NMR investigations of cellulose structure and interactions with matrix polysaccharides in plant primary cell walls. J. Exp. Bot. 2016, 67, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Keegstra, K.; Talmadge, K.W.; Bauer, W.D.; Albersheim, P. The Structure of Plant Cell Walls: III. A Model of the Walls of Suspension-cultured Sycamore Cells Based on the Interconnections of the Macromolecular Components. Plant Physiol. 1973, 51, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Wolfgang, W. Pattern formation in the monocot embryo as revealed by NAM and CUC3 orthologues from Zea mays L. Plant Mol. Biol. 2005, 58, 669–685. [Google Scholar] [CrossRef] [PubMed]
- Samaj, J.; Samajova, O.; Peters, M.; Baluska, F.; Lichtscheidl, I.; Knox, J.P.; Volkmann, D. Immunolocalization of LM2 arabinogalactan protein epitope associated with endomembranes of plant cells. Protoplasma 2000, 212, 186–196. [Google Scholar] [CrossRef]
- Bourquin, V.; Nishikubo, N.; Abe, H.; Brumer, H.; Denman, S.; Eklund, M.; Christiernin, M.; Teeri, T.T.; Sundberg, B.; Mellerowicz, E.J. Xyloglucan endotransglycosylases have a function during the formation of secondary cell walls of vascular tissues. Plant Cell 2002, 14, 3073–3088. [Google Scholar] [CrossRef] [PubMed]
- Betekhtin, A.; Rojek, M.; Milewska-Hendel, A.; Gawecki, R.; Karcz, J.; Kurczynska, E.; Hasterok, R. Spatial Distribution of Selected Chemical Cell Wall Components in the Embryogenic Callus of Brachypodium distachyon. PLoS ONE 2016, 11, e0167426. [Google Scholar] [CrossRef] [PubMed]
- Sechet, J.; Frey, A.; Effroy-Cuzzi, D.; Berger, A.; Perreau, F.; Cueff, G.; Charif, D.; Rajjou, L.; Mouille, G.; North, H.M.; et al. Xyloglucan Metabolism Differentially Impacts the Cell Wall Characteristics of the Endosperm and Embryo during Arabidopsis Seed Germination. Plant Physiol. 2016, 170, 1367–1380. [Google Scholar] [CrossRef] [PubMed]
- Ordaz-Ortiz, J.J.; Marcus, S.E.; Knox, J.P. Cell wall microstructure analysis implicates hemicellulose polysaccharides in cell adhesion in tomato fruit pericarp parenchyma. Mol. Plant 2009, 2, 910–921. [Google Scholar] [CrossRef] [PubMed]
- Cantu-Jungles, T.M.; Iacomini, M.; Cipriani, T.R.; Cordeiro, L.M.C. Extraction and characterization of pectins from primary cell walls of edible açaí (Euterpe oleraceae) berries, fruits of a monocotyledon palm. Carbohydr. Polym. 2017, 158, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Brennan, M.; Harris, P.J. Distribution of fucosylated xyloglucans among the walls of different cell types in monocotyledons determined by immunofluorescence microscopy. Mol. Plant 2011, 4, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Francin-Allami, M.; Lollier, V.; Pavlovic, M.; San Clemente, H.; Rogniaux, H.; Jamet, E.; Guillon, F.; Larre, C. Understanding the remodelling of cell walls during Brachypodium distachyon grain development through a sub-cellular quantitative proteomic approach. Proteomes 2016, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Skepper, J.N.; Powell, J.M. Ultrastructural immunochemistry. CSH Protoc. 2008, 2008. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.J.; Wood, E.A.; Larkins, A.P.; Galfre, G.; Butcher, G.W.; Brewin, N.J. Isolation of monoclonal antibodies reacting with peribacteriod membranes and other components of pea root nodules containing Rhizobium leguminosarum. Planta 1988, 173, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Pennell, R.I.; Knox, J.P.; Scofield, G.N.; Selvendran, R.; Roberts, K. A family of abundant plasma membrane-associated glycoproteins related to the arabinogalactan proteins is unique to flowering plants. J. Cell Biol. 1989, 108, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Yates, E.A.; Knox, J.P. Investigations into the occurrence of plant cell surface epitopes in exudate gums. Carbohydr. Polym. 1994, 24, 281–286. [Google Scholar] [CrossRef]
- Yates, E.A.; Valdor, J.-F.; Haslam, S.M.; Morris, H.R.; Dell, A.; Mackie, W.; Knox, J.P. Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 1996, 6, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Pennell, R.I.; Janniche, L.; Kjellbom, P.; Scofield, G.N.; Peart, J.M.; Roberts, K. Developmental regulation of a plasma membrane arabinogalactan protein epitope in oilseed rape flowers. Plant Cell 1991, 3, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Knox, J.P.; Linstead, P.J.; Peart, J.; Cooper, C.; Roberts, K. Developmentally regulated epitopes of cell surface arabinogalactan proteins and their relation to root tissue pattern formation. Plant J. 1991, 1, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, M.; Yates, E.A.; Willats, W.G.T.; Martin, H.; Knox, J.P. Immunochemical comparison of membrane-associated and secreted arabinogalactan-proteins in rice and carrot. Planta 1996, 198, 452–459. [Google Scholar] [CrossRef]
- Smallwood, M.; Martin, H.; Knox, J.P. An epitope of rice threonine-rich and hydroxyproline-rich glycoprotein is common to cell-wall and hydrophobic plasma-membrane glycoproteins. Planta 1995, 196, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, M.; Beven, A.; Donovan, N.; Neill, S.J.; Peart, J.; Roberts, K.; Knox, J.P. Localization of cell wall proteins in relation to the developmental anatomy of the carrot root apex. Plant J. 1994, 5, 237–246. [Google Scholar] [CrossRef]
- Moller, I.; Marcus, S.E.; Haeger, A.; Verhertbruggen, Y.; Verhoef, R.; Schols, H.; Ulvskov, P.; Mikkelsen, J.D.; Knox, J.P.; Willats, W. High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconj. J. 2008, 25, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Willats, W.G.; Marcus, S.E.; Knox, J.P. Generation of monoclonal antibody specific to (1-->5)-alpha-l-arabinan. Carbohydr. Res. 1998, 308, 149–152. [Google Scholar] [CrossRef]
- Marcus, S.E.; Blake, A.W.; Benians, T.A.; Lee, K.J.; Poyser, C.; Donaldson, L.; Leroux, O.; Rogowski, A.; Petersen, H.L.; Boraston, A.; et al. Restricted access of proteins to mannan polysaccharides in intact plant cell walls. Plant J. 2010, 64, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, H.L.; Fangel, J.U.; McCleary, B.; Ruzanski, C.; Rydahl, M.G.; Ralet, M.C.; Farkas, V.; von Schantz, L.; Marcus, S.E.; Andersen, M.C.; et al. Versatile high resolution oligosaccharide microarrays for plant glycobiology and cell wall research. J. Biol. Chem. 2012, 287, 39429–39438. [Google Scholar] [CrossRef] [PubMed]
- Luft, J.H. Improvements in epoxy resin embedding methods. J. Biophys. Biochem. Cytol. 1961, 9, 409–414. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, D.L.; Elton, S.; Ticchiarelli, F.; Hsia, M.M.; Vogel, J.P.; Leyser, O. Cross-species functional diversity within the PIN auxin efflux protein family. elife 2017, 6, e31804. [Google Scholar] [CrossRef] [PubMed]









| Antibody | Epitope | References |
|---|---|---|
| Arabinogalactan proteins (AGPs) | ||
| MAC207 | β-GlcA1->3αGalA1->2Rha | [53,54,55,56] |
| JIM8 | Arabinogalactan | [57] |
| JIM13 | βGlcA1->3αGalA1->2Rha | [55,56,58] |
| JIM16 | AGP glycan | [55,56,58] |
| LM2 | β-linked GlcA | [56,59] |
| Extensins | ||
| LM1 | Extensin | [60] |
| JIM11 | Extensin | [55,61] |
| JIM12 | Extensin | [61] |
| Pectins | ||
| LM19 | α-GalA(1-4)α-GalA(1-4)α-GalA(1-4)α-GalA | [35] |
| LM13 | α-1,5-arabinan | [62] |
| LM16 | Processed arabinan—rhamnogalacturonan (RG)-I domain | [35] |
| LM6 | αAra1-5αAra1-5αAra1-5αAra1-5Ara | [63] |
| Hemicelluloses | ||
| LM21 | β-linked mannan polysaccharides of plant cell walls | [64] |
| LM25 | Xyloglucan | [65] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betekhtin, A.; Milewska-Hendel, A.; Lusinska, J.; Chajec, L.; Kurczynska, E.; Hasterok, R. Organ and Tissue-Specific Localisation of Selected Cell Wall Epitopes in the Zygotic Embryo of Brachypodium distachyon. Int. J. Mol. Sci. 2018, 19, 725. https://doi.org/10.3390/ijms19030725
Betekhtin A, Milewska-Hendel A, Lusinska J, Chajec L, Kurczynska E, Hasterok R. Organ and Tissue-Specific Localisation of Selected Cell Wall Epitopes in the Zygotic Embryo of Brachypodium distachyon. International Journal of Molecular Sciences. 2018; 19(3):725. https://doi.org/10.3390/ijms19030725
Chicago/Turabian StyleBetekhtin, Alexander, Anna Milewska-Hendel, Joanna Lusinska, Lukasz Chajec, Ewa Kurczynska, and Robert Hasterok. 2018. "Organ and Tissue-Specific Localisation of Selected Cell Wall Epitopes in the Zygotic Embryo of Brachypodium distachyon" International Journal of Molecular Sciences 19, no. 3: 725. https://doi.org/10.3390/ijms19030725
APA StyleBetekhtin, A., Milewska-Hendel, A., Lusinska, J., Chajec, L., Kurczynska, E., & Hasterok, R. (2018). Organ and Tissue-Specific Localisation of Selected Cell Wall Epitopes in the Zygotic Embryo of Brachypodium distachyon. International Journal of Molecular Sciences, 19(3), 725. https://doi.org/10.3390/ijms19030725