Upregulation of FLG, LOR, and IVL Expression by Rhodiola crenulata Root Extract via Aryl Hydrocarbon Receptor: Differential Involvement of OVOL1
Abstract
:1. Introduction
2. Results
2.1. RCE Is an AHR Agonist
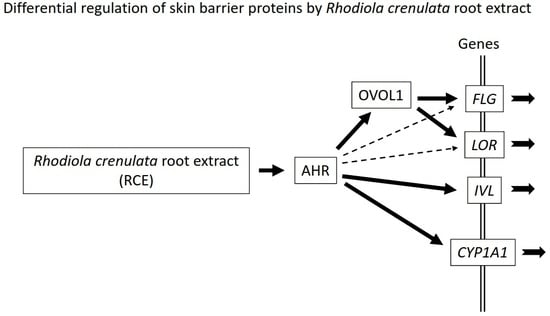
2.2. RCE Upregulates FLG, LOR, and IVL Expression in an AHR-Dependent Fashion
2.3. RCE Upregulates OVOL1 Expression in an AHR-Dependent Manner
2.4. RCE-Mediated IVL Upregulation Is OVOL1-Independent
3. Discussion
3.1. Barrier Function and AHR-OVOL1 Axis
3.2. Upregulation of FLG, LOR, and IVL by AHR Activation
3.3. Differential Regulation of FLG, LOR, and INV by AHR-OVOL1 Signaling
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Cell Culture
4.3. Immunofluorescence and Confocal Laser Scanning Microscopic Analysis
4.4. Reverse Transcription-PCR and qRT-PCR Analyses
4.5. Transfection with siRNAs against AHR and OVOL1
4.6. Western Blot Analysis
4.7. Statistical Analysis
Supplementary Materials
Author contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| AHR | Aryl hydrocarbon receptor |
| CYP1A1 | Cytochrome P450 1A1 |
| FLG | Filaggrin |
| IVL | Involucrin |
| LOR | Loricrin |
| NHEK | Normal human epidermal keratinocyte |
| OVOL1 | OVO-like 1 |
| RCE | Rhodiola crenulata root extract |
References
- Kypriotou, M.; Huber, M.; Hohl, D. The human epidermal differentiation complex: Cornified envelope precursors, S100 proteins and the ‘fused genes’ family. Exp. Dermatol. 2012, 21, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Esser, C.; Bargen, I.; Weighardt, H.; Haarmann-Stemmann, T.; Krutmann, J. Functions of the aryl hydrocarbon receptor in the skin. Semin. Immunopathol. 2013, 35, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Furue, M.; Takahara, M.; Nakahara, T.; Uchi, H. Role of AhR/ARNT system in skin homeostasis. Arch. Dermatol. Res. 2014, 306, 769–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furue, M.; Uchi, H.; Mitoma, C.; Hashimoto-Hachiya, A.; Chiba, T.; Ito, T.; Nakahara, T.; Tsuji, G. Antioxidants for healthy skin: The emerging role of aryl hydrocarbon receptors and nuclear factor-erythroid 2-related factor-2. Nutrients 2017, 9, 223. [Google Scholar] [CrossRef] [PubMed]
- Sutter, C.H.; Bodreddigari, S.; Campion, C.; Wible, R.S.; Sutter, T.R. 2,3,7,8-Tetrachlorodibenzo-p-dioxin increases the expression of genes in the human epidermal differentiation complex and accelerates epidermal barrier formation. Toxicol. Sci. 2011, 124, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Furue, M.; Tsuji, G.; Mitoma, C.; Nakahara, T.; Chiba, T.; Morino-Koga, S.; Uchi, H. Gene regulation of filaggrin and other skin barrier proteins via aryl hydrocarbon receptor. J. Dermatol. Sci. 2015, 80, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Mitoma, C.; Hashimoto-Hachiya, A.; Uchi, H.; Takahara, M.; Tsuji, G.; Kido-Nakahara, M.; Nakahara, T.; Furue, M. Antioxidant soybean tar Glyteer rescues T-helper-mediated downregulation of filaggrin expression via aryl hydrocarbon receptor. J. Dermatol. 2015, 42, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Haas, K.; Weighardt, H.; Deenen, R.; Köhrer, K.; Clausen, B.; Zahner, S.; Boukamp, P.; Bloch, W.; Krutmann, J.; Esser, C. Aryl hydrocarbon receptor in keratinocytes is essential for murine skin barrier integrity. J. Investig. Dermatol. 2016, 136, 2260–2269. [Google Scholar] [CrossRef] [PubMed]
- Van den Bogaard, E.H.; Bergboer, J.G.; Vonk-Bergers, M.; van Vlijmen-Willems, I.M.; Hato, S.V.; van der Valk, P.G.; Schröder, J.M.; Joosten, I.; Zeeuwen, P.L.; Schalkwijk, J. Coal tar induces AHR-dependent skin barrier repair in atopic dermatitis. J. Clin. Investig. 2013, 123, 917–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takei, K.; Mitoma, C.; Hashimoto-Hachiya, A.; Takahara, M.; Tsuji, G.; Nakahara, T.; Furue, M. Galactomyces fermentation filtrate prevents T helper 2-mediated reduction of filaggrin in an aryl hydrocarbon receptor-dependent manner. Clin. Exp. Dermatol. 2015, 40, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, G.; Hashimoto-Hachiya, A.; Kiyomatsu-Oda, M.; Takemura, M.; Ohno, F.; Ito, T.; Morino-Koga, S.; Mitoma, C.; Nakahara, T.; Uchi, H.; et al. Aryl hydrocarbon receptor activation restores filaggrin expression via OVOL1 in atopic dermatitis. Cell Death Dis. 2017, 8, e2931. [Google Scholar] [CrossRef] [PubMed]
- Hirano, A.; Goto, M.; Mitsui, T.; Hashimoto-Hachiya, A.; Tsuji, G.; Furue, M. Antioxidant Artemisia princeps extract enhances the expression of filaggrin and loricrin via the AHR/OVOL1 pathway. Int. J. Mol. Sci. 2017, 18, 1948. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Schonbaum, C.; Degenstein, L.; Bai, W.; Mahowald, A.; Fuchs, E. The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev. 1998, 12, 3452–3463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, T.; Tsuji, G.; Ohno, F.; Uchi, H.; Nakahara, T.; Hashimoto-Hachiya, A.; Yoshida, Y.; Yamamoto, O.; Oda, Y.; Furue, M. Activation of the OVOL1-OVOL2 axis in the hair bulb and in pilomatricoma. Am. J. Pathol. 2016, 186, 1036–1043. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Tsuji, G.; Ohno, F.; Nakahara, T.; Uchi, H.; Furue, M. Potential role of the OVOL1-OVOL2 axis and c-Myc in the progression of cutaneous squamous cell carcinoma. Mod. Pathol. 2017, 30, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Nair, M.; Teng, A.; Bilanchone, V.; Agrawal, A.; Li, B.; Dai, X. Ovol1 regulates the growth arrest of embryonic epidermal progenitor cells and represses c-myc transcription. J. Cell Biol. 2006, 173, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.T.; Chang, T.C.; Lai, F.Y.; Lin, C.S.; Chao, H.L.; Lee, S.Y. Rhodiola crenulata attenuates γ-ray induced cellular injury via modulation of oxidative stress in human skin cells. Am. J. Chin. Med. 2018, 46, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.Y.; Pang, X.W.; Zhang, G.Q.; Guo, J.Y. Salidroside’s protection against UVB-mediated oxidative damage and apoptosis is associated with the upregulation of Nrf2 expression. Photomed. Laser Surg. 2017, 35, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Li, Y.; Ma, L.; Liu, T.; Wu, Y.; Xu, R.; Song, A.; Yin, R. A rapid and sensitiveUHPLC-FT-ICR MS/MS method for identification of chemical constituents in Rhodiola crenulata extract, rat plasm and rat brain after oral administration. Talanta 2016, 160, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, L.H.; Sutter, C.H.; Leon Carrion, S.; Tran, Q.T.; Bodreddigari, S.; Kensicki, E.; Mohney, R.P.; Sutter, T.R. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-mediated production of reactive oxygen species is an essential step in the mechanism of action to accelerate human keratinocyte differentiation. Toxicol. Sci. 2013, 132, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Murray, I.A.; Nichols, R.G.; Zhang, L.; Patterson, A.D.; Perdew, G.H. Expression of the aryl hydrocarbon receptor contributes to the establishment of intestinal microbial community structure in mice. Sci. Rep. 2016, 6, 33969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goettel, J.A.; Gandhi, R.; Kenison, J.E.; Yeste, A.; Murugaiyan, G.; Sambanthamoorthy, S.; Griffith, A.E.; Patel, B.; Shouval, D.S.; Weiner, H.L.; et al. AHR activation is protective against colitis driven by T cells in humanized mice. Cell Rep. 2016, 17, 1318–1329. [Google Scholar] [CrossRef] [PubMed]
- Chiba, T.; Chihara, J.; Furue, M. Role of the arylhydrocarbon receptor (AhR) in the pathology of asthma and COPD. J. Allergy 2012, 2012, 372384. [Google Scholar] [CrossRef] [PubMed]
- Chiba, T.; Uchi, H.; Yasukawa, F.; Furue, M. Role of the arylhydrocarbon receptor in lung disease. Int. Arch. Allergy Immunol. 2011, 155 (Suppl. 1), 129–134. [Google Scholar] [CrossRef] [PubMed]
- Chiba, T.; Uchi, H.; Tsuji, G.; Gondo, H.; Moroi, Y.; Furue, M. Arylhydrocarbon receptor (AhR) activation in airway epithelial cells induces MUC5AC via reactive oxygen species (ROS) production. Pulm. Pharmacol. Ther. 2011, 24, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Gaitanis, G.; Magiatis, P.; Stathopoulou, K.; Bassukas, I.D.; Alexopoulos, E.C.; Velegraki, A.; Skaltsounis, A.L. AhR ligands, malassezin, and indolo[3,2-b]carbazole are selectively produced by Malassezia furfur strains isolated from seborrheic dermatitis. J. Investig. Dermatol. 2008, 128, 1620–1625. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, E.; Schäfer, C.; Calles, C.; Bernsmann, T.; Bernshausen, T.; Wurm, M.; Hübenthal, U.; Cline, J.E.; Hajimiragha, H.; Schroeder, P.; et al. Lightening up the UV response by identification of the arylhydrocarbon receptor as a cytoplasmatic target for ultraviolet B radiation. Proc. Natl. Acad. Sci. USA 2007, 104, 8851–8856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, M.; Man, M.; Man, W.; Zhu, W.; Hupe, M.; Park, K.; Crumrine, D.; Elias, P.M.; Man, M.Q. Topical hesperidin improves epidermal permeability barrier function and epidermal differentiation in normal murine skin. Exp. Dermatol. 2012, 21, 337–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, M.; Sun, R.; Hupe, M.; Kim, P.L.; Park, K.; Crumrine, D.; Lin, T.K.; Santiago, J.L.; Mauro, T.M.; Elias, P.M.; et al. Topical apigenin improves epidermal permeability barrier homoeostasis in normal murine skin by divergent mechanisms. Exp. Dermatol. 2013, 22, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.N.; Irvine, A.D.; Terron-Kwiatkowski, A.; Zhao, Y.; Liao, H.; Lee, S.P.; Goudie, D.R.; Sandilands, A.; Campbell, L.E.; Smith, F.J.; et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 2006, 38, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Esparza-Gordillo, J.; Weidinger, S.; Fölster-Holst, R.; Bauerfeind, A.; Ruschendorf, F.; Patone, G.; Rohde, K.; Marenholz, I.; Schulz, F.; Kerscher, T.; et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat. Genet. 2009, 41, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.D.; Xiao, F.L.; Li, Y.; Zhou, W.M.; Tang, H.Y.; Tang, X.F.; Zhang, H.; Schaarschmidt, H.; Zuo, X.B.; Foelster-Holst, R.; et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat. Genet. 2011, 43, 690–694. [Google Scholar] [CrossRef] [PubMed]
- Paternoster, L.; Standl, M.; Chen, C.M.; Ramasamy, A.; Bønnelykke, K.; Duijts, L.; Ferreira, M.A.; Alves, A.C.; Thyssen, J.P.; Albrecht, E.; et al. Meta-analysis of genome-wide association studies identifies three new risk loci for atopic dermatitis. Nat. Genet. 2011, 44, 187–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirota, T.; Takahashi, A.; Kubo, M.; Tsunoda, T.; Tomita, K.; Sakashita, M.; Yamada, T.; Fujieda, S.; Tanaka, S.; Doi, S.; et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat. Genet. 2012, 44, 1222–1226. [Google Scholar] [CrossRef] [PubMed]
- Tamari, M.; Hirota, T. Genome-wide association studies of atopic dermatitis. J. Dermatol. 2014, 41, 213–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paternoster, L.; Standl, M.; Waage, J.; Baurecht, H.; Hotze, M.; Strachan, D.P.; Curtin, J.A.; Bønnelykke, K.; Tian, C.; Takahashi, A.; et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat. Genet. 2015, 47, 1449–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seguchi, T.; Cui, C.Y.; Kusuda, S.; Takahashi, M.; Aisu, K.; Tezuka, T. Decreased expression of filaggrin in atopic skin. Arch. Dermatol. Res. 1996, 288, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Guttman-Yassky, E.; Ungar, B.; Malik, K.; Dickstein, D.; Suprun, M.; Estrada, Y.D.; Xu, H.; Peng, X.; Oliva, M.; Todd, D.; et al. Molecular signatures order the potency of topically applied anti-inflammatory drugs in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2017, 140, 1032–1042. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.M.; Scherer, A.; Wanke, C.; Bräutigam, M.; Bongiovanni, S.; Letzkus, M.; Staedtler, F.; Kehren, J.; Zuehlsdorf, M.; Schwarz, T.; et al. Gene expression is differently affected by pimecrolimus and betamethasone in lesional skin of atopic dermatitis. Allergy 2012, 67, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Furue, M.; Chiba, T.; Tsuji, G.; Ulzii, D.; Kido-Nakahara, M.; Nakahara, T.; Kadono, T. Atopic dermatitis: Immune deviation, barrier dysfunction, IgE autoreactivity and new therapies. Allergol. Int. 2017, 66, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Bissonnette, R.; Poulin, Y.; Zhou, Y.; Tan, J.; Hong, H.C.; Webster, J.; Ip, W.; Tang, L.; Lyle, M. Efficacy and safety of topical WBI-1001 in patients with mild to severe atopic dermatitis: Results from a 12-week, multicentre, randomized, placebo-controlled double-blind trial. Br. J. Dermatol. 2012, 166, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Bissonnette, R.; Bolduc, C.; Maari, C.; Nigen, S.; Webster, J.M.; Tang, L.; Lyle, M. Efficacy and safety of topical WBI-1001 in patients with mild to moderate psoriasis: Results from a randomized double-blind placebo-controlled, phase II trial. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 1516–1521. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.H.; Jayawickreme, C.; Rickard, D.J.; Nicodeme, E.; Bui, T.; Simmons, C.; Coquery, C.M.; Neil, J.; Pryor, W.M.; Mayhew, D.; et al. Tapinarof is a natural AhR agonist that resolves skin inflammation in mice and humans. J. Investig. Dermatol. 2017, 137, 2110–2119. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashimoto-Hachiya, A.; Tsuji, G.; Murai, M.; Yan, X.; Furue, M. Upregulation of FLG, LOR, and IVL Expression by Rhodiola crenulata Root Extract via Aryl Hydrocarbon Receptor: Differential Involvement of OVOL1. Int. J. Mol. Sci. 2018, 19, 1654. https://doi.org/10.3390/ijms19061654
Hashimoto-Hachiya A, Tsuji G, Murai M, Yan X, Furue M. Upregulation of FLG, LOR, and IVL Expression by Rhodiola crenulata Root Extract via Aryl Hydrocarbon Receptor: Differential Involvement of OVOL1. International Journal of Molecular Sciences. 2018; 19(6):1654. https://doi.org/10.3390/ijms19061654
Chicago/Turabian StyleHashimoto-Hachiya, Akiko, Gaku Tsuji, Mika Murai, Xianghong Yan, and Masutaka Furue. 2018. "Upregulation of FLG, LOR, and IVL Expression by Rhodiola crenulata Root Extract via Aryl Hydrocarbon Receptor: Differential Involvement of OVOL1" International Journal of Molecular Sciences 19, no. 6: 1654. https://doi.org/10.3390/ijms19061654
APA StyleHashimoto-Hachiya, A., Tsuji, G., Murai, M., Yan, X., & Furue, M. (2018). Upregulation of FLG, LOR, and IVL Expression by Rhodiola crenulata Root Extract via Aryl Hydrocarbon Receptor: Differential Involvement of OVOL1. International Journal of Molecular Sciences, 19(6), 1654. https://doi.org/10.3390/ijms19061654