Calcium-Binding Proteins S100A8 and S100A9: Investigation of Their Immune Regulatory Effect in Myeloid Cells
Abstract
:1. Introduction
2. Results
2.1. S100A9 is Mostly Expressed in CD14-Positive (Classical) Monocytes
2.2. S100A9 Expression Varies within the CD14+ Monocyte Population
2.3. Both HLA-D-Positive Monocytes and Myeloid Derived Suppressor Cells Express S100A9
2.4. Phenotypic Characterization of Cytokine Expression Profiles in S100A9high and S100A9low Monocytes
2.5. Sorting of Viable Cells: Application of SmartFlare
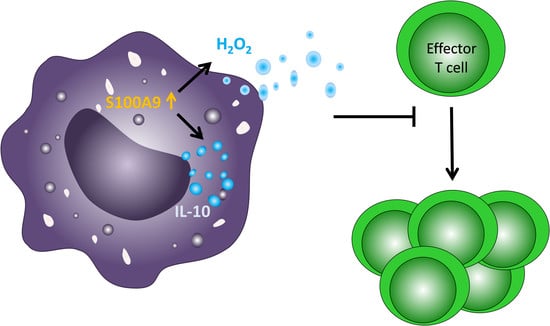
2.6. Overexpression of S100A8 and S100A9 in Monocyte-Derived Macrophages Leads to Increased ROS Production and Elevated IL-10 mRNA Expression
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Monocyte Purification
4.3. Monocyte Subsets FACS Sorting
4.4. Measurement of S100A9 in Monocytes Using the Cytospin Method
4.5. Intracellular Staining and FACS Sorting
4.6. RNA Isolation and cDNA Synthesis
4.7. Real-Time Quantitative PCR
4.8. Overexpression of S100A8/A9 and Measurement of ROS and Cytokines
4.9. Statistical Analyses
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Kane, D.; Roth, J.; Frosch, M.; Vogl, T.; Bresnihan, B.; FitzGerald, O. Increased perivascular synovial membrane expression of myeloid-related proteins in psoriatic arthritis. Arthritis Rheumatol. 2003, 48, 1676–1685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odink, K.; Cerletti, N.; Brüggen, J.; Clerc, R.G.; Tarcsay, L.; Zwadlo, G.; Gerhards, G.; Schlegel, R.; Sorg, C. Two calcium-binding proteins in infiltrate macrophages of rheumatoid arthritis. Nature 1987, 330, 80–82. [Google Scholar] [CrossRef] [PubMed]
- Ryckman, C.; Vandal, K.; Rouleau, P.; Talbot, M.; Tessier, P.A. Proinflammatory activities of S100: Proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J. Immunol. 2003, 170, 3233–3242. [Google Scholar] [CrossRef] [PubMed]
- Pechkovsky, D.; Zalutskaya, O.; Ivanov, G.; Misuno, N. Calprotectin (MRP8/14 protein complex) release during mycobacterial infection in vitro and in vivo. FEMS Immunol. Med. Microbiol. 2000, 29, 27–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frosch, M.; Strey, A.; Vogl, T.; Wulffraat, N.M.; Kuis, W.; Sunderkötter, C.; Harms, E.; Sorg, C.; Roth, J. Myeloid-related proteins 8 and 14 are specifically secreted during interaction of phagocytes and activated endothelium and are useful markers for monitoring disease activity in pauciarticular-onset juvenile rheumatoid arthritis. Arthritis Rheumatol. 2000, 43, 628–637. [Google Scholar] [CrossRef] [Green Version]
- Leukert, N.; Vogl, T.; Strupat, K.; Reichelt, R.; Sorg, C.; Roth, J. Calcium-dependent tetramer formation of S100A8 and S100A9 is essential for biological activity. J. Mol. Biol. 2006, 359, 961–972. [Google Scholar] [CrossRef] [PubMed]
- Murthy, A.; Lehrer, R.; Harwig, S.; Miyasaki, K. In vitro candidastatic properties of the human neutrophil calprotectin complex. J. Immunol. 1993, 151, 6291–6301. [Google Scholar] [PubMed]
- Sohnle, P.G.; Hunter, M.J.; Hahn, B.; Chazin, W.J. Zinc-reversible antimicrobial activity of recombinant calprotectin (migration inhibitory factor—Related proteins 8 and 14). J. Infect. Dis. 2000, 182, 1272–1275. [Google Scholar] [CrossRef] [PubMed]
- Vogl, T.; Tenbrock, K.; Ludwig, S.; Leukert, N.; Ehrhardt, C.; Van Zoelen, M.A.; Nacken, W.; Foell, D.; Van der Poll, T.; Sorg, C. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat. Med. 2007, 13, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Boyd, J.H.; Kan, B.; Roberts, H.; Wang, Y.; Walley, K.R. S100A8 and S100A9 mediate endotoxin-induced cardiomyocyte dysfunction via the receptor for advanced glycation end products. Circ. Res. 2008, 102, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, K.; Terasaki, F.; Ikemoto, M.; Fujita, S.; Tsukada, B.; Katashima, T.; Kanzaki, Y.; Sohmiya, K.; Kono, T.; Toko, H. Suppression of inflammation in rat autoimmune myocarditis by S100A8/A9 through modulation of the proinflammatory cytokine network. Eur. J. Heart Fail. 2009, 11, 229–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikemoto, M.; Murayama, H.; Itoh, H.; Totani, M.; Fujita, M. Intrinsic function of S100A8/A9 complex as an anti-inflammatory protein in liver injury induced by lipopolysaccharide in rats. Clin. Chim. Acta 2007, 376, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, P.; Corzo, C.A.; Luetteke, N.; Yu, B.; Nagaraj, S.; Bui, M.M.; Ortiz, M.; Nacken, W.; Sorg, C.; Vogl, T. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J. Exp. Med. 2008, 205, 2235–2249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinha, P.; Okoro, C.; Foell, D.; Freeze, H.H.; Ostrand-Rosenberg, S.; Srikrishna, G. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J. Immunol. 2008, 181, 4666–4675. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Hoechst, B.; Duffy, A.; Gamrekelashvili, J.; Fioravanti, S.; Manns, M.P.; Greten, T.F.; Korangy, F. S100A9 a new marker for monocytic human myeloid-derived suppressor cells. Immunology 2012, 136, 176–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuidwijk, K.; de Fijter, J.W.; Mallat, M.J.; Eikmans, M.; van Groningen, M.C.; Goemaere, N.N.; Bajema, I.M.; van Kooten, C. Increased influx of myeloid dendritic cells during acute rejection is associated with interstitial fibrosis and tubular atrophy and predicts poor outcome. Kidney Int. 2012, 81, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Toki, D.; Zhang, W.; Hor, K.; Liuwantara, D.; Alexander, S.; Yi, Z.; Sharma, R.; Chapman, J.; Nankivell, B.; Murphy, B. The role of macrophages in the development of human renal allograft fibrosis in the first year after transplantation. Am. J. Transplant. 2014, 14, 2126–2136. [Google Scholar] [CrossRef] [PubMed]
- Bergler, T.; Jung, B.; Bourier, F.; Kühne, L.; Banas, M.C.; Rümmele, P.; Wurm, S.; Banas, B. Infiltration of macrophages correlates with severity of allograft rejection and outcome in human kidney transplantation. PLoS ONE 2016, 11, e0156900. [Google Scholar] [CrossRef] [PubMed]
- Eikmans, M.; Roos-van Groningen, M.C.; Sijpkens, Y.W.; Ehrchen, J.; Roth, J.; Baelde, H.J.; Bajema, I.M.; de Fijter, J.W.; de Heer, E.; Bruijn, J.A. Expression of surfactant protein-C, S100A8, S100A9, and B cell markers in renal allografts: Investigation of the prognostic value. J. Am. Soc. Nephrol. 2005, 16, 3771–3786. [Google Scholar] [CrossRef] [PubMed]
- Rekers, N.V.; Bajema, I.M.; Mallat, M.J.; Petersen, B.; Anholts, J.D.; Swings, G.M.; van Miert, P.P.; Kerkhoff, C.; Roth, J.; Popp, D.; et al. Beneficial Immune Effects of Myeloid-Related Proteins in Kidney Transplant Rejection. Am. J. Transplant. 2016, 16, 1441–1455. [Google Scholar] [CrossRef] [PubMed]
- Bronte, V.; Brandau, S.; Chen, S.H.; Colombo, M.P.; Frey, A.B.; Greten, T.F.; Mandruzzato, S.; Murray, P.J.; Ochoa, A.; Ostrand-Rosenberg, S.; et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016, 7, 12150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, Y.; Mosheir, E.; Menon, M.; Wilson, D.; Woytovich, C.; Ochando, J.; Murphy, B. Monocytic Myeloid-Derived Suppressor Cells Accumulate in Renal Transplant Patients and Mediate CD4+ Foxp3+ Treg Expansion. Am. J. Transplant. 2013, 13, 3123–3131. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Chen, S.; Guo, X.; Chen, Z.; Huang, X.; Lai, Y.; Lin, M. Clinical significance of myeloid-derived suppressor cells in human renal transplantation with acute T cell-mediated rejection. Inflammation 2014, 37, 1799–1805. [Google Scholar] [CrossRef] [PubMed]
- Boyette, L.B.; Macedo, C.; Hadi, K.; Elinoff, B.D.; Walters, J.T.; Ramaswami, B.; Chalasani, G.; Taboas, J.M.; Lakkis, F.G.; Metes, D.M. Phenotype, function, and differentiation potential of human monocyte subsets. PLoS ONE 2017, 12, e0176460. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.L.; Tai, J.J.-Y.; Wong, W.-C.; Han, H.; Sem, X.; Yeap, W.-H.; Kourilsky, P.; Wong, S.-C. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 2011, 118, e16–e31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziegler-Heitbrock, L.; Ancuta, P.; Crowe, S.; Dalod, M.; Grau, V.; Hart, D.N.; Leenen, P.J.; Liu, Y.-J.; MacPherson, G.; Randolph, G.J. Nomenclature of monocytes and dendritic cells in blood. Blood 2010, 116, e74–e80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ancuta, P.; Liu, K.-Y.; Misra, V.; Wacleche, V.S.; Gosselin, A.; Zhou, X.; Gabuzda, D. Transcriptional profiling reveals developmental relationship and distinct biological functions of CD16+ and CD16-monocyte subsets. BMC Genom. 2009, 10, 403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Zhang, H.; Wong, W.-C.; Sem, X.; Han, H.; Ong, S.-M.; Tan, Y.-C.; Yeap, W.-H.; Gan, C.-S.; Ng, K.-Q. Identification of novel functional differences in monocyte subsets using proteomic and transcriptomic methods. J. Proteome Res. 2009, 8, 4028–4038. [Google Scholar] [CrossRef] [PubMed]
- Hrvatin, S.; Deng, F.; O’Donnell, C.W.; Gifford, D.K.; Melton, D.A. MARIS: Method for analyzing RNA following intracellular sorting. PLoS ONE 2014, 9, e89459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, A.I.; Wiltshire, T.; Batalov, S.; Lapp, H.; Ching, K.A.; Block, D.; Zhang, J.; Soden, R.; Hayakawa, M.; Kreiman, G. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 2004, 101, 6062–6067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prigodich, A.E.; Seferos, D.S.; Massich, M.D.; Giljohann, D.A.; Lane, B.C.; Mirkin, C.A. Nano-flares for mRNA regulation and detection. ACS Nano 2009, 3, 2147–2152. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Menzel, U.; Loebel, C.; Schmal, H.; Alini, M.; Stoddart, M.J. Monitoring live human mesenchymal stromal cell differentiation and subsequent selection using fluorescent RNA-based probes. Sci. Rep. 2016, 6, 26014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halo, T.L.; McMahon, K.M.; Angeloni, N.L.; Xu, Y.; Wang, W.; Chinen, A.B.; Malin, D.; Strekalova, E.; Cryns, V.L.; Cheng, C. NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc. Natl. Acad. Sci. USA 2014, 111, 17104–17109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seferos, D.S.; Giljohann, D.A.; Hill, H.D.; Prigodich, A.E.; Mirkin, C.A. Nano-flares: Probes for transfection and mRNA detection in living cells. J. Am. Chem. Soc. 2007, 129, 15477–15479. [Google Scholar] [CrossRef] [PubMed]
- Czarnek, M.; Bereta, J. SmartFlares fail to reflect their target transcripts levels. Sci. Rep. 2017, 7, 11682. [Google Scholar] [CrossRef] [PubMed]
- Levy, R.; Held, M.; Mason, D.; Comenge, J.; Carolan, G. The spherical nucleic acids mRNA detection paradox. ScienceOpen Res. 2015. [Google Scholar] [CrossRef]
- Wu, X.A.; Choi, C.H.J.; Zhang, C.; Hao, L.; Mirkin, C.A. Intracellular fate of spherical nucleic acid nanoparticle conjugates. J. Am. Chem. Soc. 2014, 136, 7726–7733. [Google Scholar] [CrossRef] [PubMed]
- Belikov, A.V.; Schraven, B.; Simeoni, L. T cells and reactive oxygen species. J. Biomed. Sci. 2015, 22, 85. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Patel, S.; Tcyganov, E.; Gabrilovich, D.I. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016, 37, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Pearce, A.; Traulsen, K.; Rintala, A.; Lee, H. Fluorescence produced by transfection reagents can be confused with green fluorescent proteins in mammalian cells. Biotechniques 2001, 31, 314–316. [Google Scholar] [PubMed]
- Yang, J.; Kemps-Mols, B.; Spruyt-Gerritse, M.; Anholts, J.; Claas, F.; Eikmans, M. The source of SYBR green master mix determines outcome of nucleic acid amplification reactions. BMC Res. Notes 2016, 9, 292. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Anholts, J.; Kolbe, U.; Stegehuis-Kamp, J.A.; Claas, F.H.J.; Eikmans, M. Calcium-Binding Proteins S100A8 and S100A9: Investigation of Their Immune Regulatory Effect in Myeloid Cells. Int. J. Mol. Sci. 2018, 19, 1833. https://doi.org/10.3390/ijms19071833
Yang J, Anholts J, Kolbe U, Stegehuis-Kamp JA, Claas FHJ, Eikmans M. Calcium-Binding Proteins S100A8 and S100A9: Investigation of Their Immune Regulatory Effect in Myeloid Cells. International Journal of Molecular Sciences. 2018; 19(7):1833. https://doi.org/10.3390/ijms19071833
Chicago/Turabian StyleYang, Jianxin, Jacqueline Anholts, Ulrike Kolbe, Janine A. Stegehuis-Kamp, Frans H. J. Claas, and Michael Eikmans. 2018. "Calcium-Binding Proteins S100A8 and S100A9: Investigation of Their Immune Regulatory Effect in Myeloid Cells" International Journal of Molecular Sciences 19, no. 7: 1833. https://doi.org/10.3390/ijms19071833
APA StyleYang, J., Anholts, J., Kolbe, U., Stegehuis-Kamp, J. A., Claas, F. H. J., & Eikmans, M. (2018). Calcium-Binding Proteins S100A8 and S100A9: Investigation of Their Immune Regulatory Effect in Myeloid Cells. International Journal of Molecular Sciences, 19(7), 1833. https://doi.org/10.3390/ijms19071833