Role of Arginine 117 in Substrate Recognition by Human Cytochrome P450 2J2
Abstract
:1. Introduction
2. Results and Discussion
2.1. Expression and Stability of CYP2J2 R117 Mutants
2.2. Oxidation of Ebastine by CYP2J2 and Its R117 Mutants
2.3. Inhibitory Effects of Terfenadone Derivatives on Ebastine Hydroxylation by CYP2J2 and Its R117 Mutants
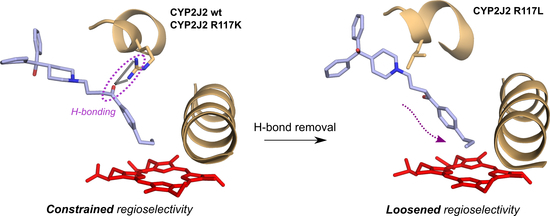
2.4. Influence of R117 Mutations on the Regioselectivity of Terfenadone Derivatives Oxidation
2.5. Homology Modeling of CYP2J2-R117X Mutants
2.6. Docking of Terfenadone Derivatives in the Active Site of CYP2J2 and Its Mutants
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Mutagenesis of the R117 Residue of the CYP2J2 cDNA
4.3. Expression of the CYP2J2 R117 Mutants
4.4. Oxidation of Terfenadone Derivatives Assay and Analysis of Product Formation Regioselectivity
4.5. Homology Modeling of CYP2J2-R117X Mutants and Docking of Derivatives 2 and 4 in CYP2J2-R117X Active Sites
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | arachidonic acid |
| CYP | cytochrome P450 |
| EET | cis-epoxyeicosatrienoic acid |
| HPLC | high performance liquid chromatography |
| MD | molecular dynamics |
| MS | mass spectrometry |
| NADP | nicotinamine adenine dinucleotide phosphate |
| NADPH | reduced nicotinamide adenine dinucleotide phosphate |
| wt | wild type |
References
- Ortiz de Montellano, P.R. Cytochrome P450: Structure, Mechanism, and Biochemistry, 4th ed.; Springer: New York, NY, USA, 2015. [Google Scholar]
- Guengerich, F.P. Human cytochrome P450 enzymes. In Cytochrome P450: Structure, Mechanism, and Biochemistry, 4th ed.; Ortiz de Montellano, P.R., Ed.; Springer: New York, NY, USA, 2015; pp. 523–786. [Google Scholar]
- Wu, S.; Moomaw, C.R.; Tomer, K.B.; Falck, J.R.; Zeldin, D.C. Molecular cloning and expression of CYP2J2, a human cytochrome P450 arachidonic acid epoxygenase highly expressed in heart. J. Biol. Chem. 1996, 271, 3460–3468. [Google Scholar] [CrossRef] [PubMed]
- Zeldin, D.C.; Foley, J.; Goldsworthy, S.M.; Cook, M.E.; Boyle, J.E.; Ma, J.; Moomaw, C.R.; Tomer, K.B.; Steenbergen, C.; Wu, S. CYP2J subfamily cytochrome P450s in the gastrointestinal tract: Expression, localization, and potential functional significance. Mol. Pharmacol. 1997, 51, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Zeldin, D.C.; Foley, J.; Ma, J.; Boyle, J.E.; Pascual, J.M.; Moomaw, C.R.; Tomer, K.B.; Steenbergen, C.; Wu, S. CYP2J subfamily P450s in the lung: Expression, localization, and potential functional significance. Mol. Pharmacol. 1996, 50, 1111–1117. [Google Scholar] [PubMed]
- Dutheil, F.; Dauchy, S.; Diry, M.; Sazdovitch, V.; Cloarec, O.; Mellottee, L.; Bieche, I.; Ingelman-Sundberg, M.; Flinois, J.P.; de Waziers, I.; et al. Xenobiotic-metabolizing enzymes and transporters in the normal human brain: Regional and cellular mapping as a basis for putative roles in cerebral function. Drug Metab. Dispos. 2009, 37, 1528–1538. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Ju, W.; Hao, H.; Wang, G.; Li, P. Cytochrome P450 2J2: Distribution, function, regulation, genetic polymorphisms and clinical significance. Drug Metab. Rev. 2013, 45, 311–352. [Google Scholar] [CrossRef] [PubMed]
- Murray, M. CYP2J2—Regulation, function and polymorphism. Drug Metab. Rev. 2016, 48, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Solanki, M.S.; Pointon, A.; Jones, B.C.; Herbert, K.E. Cytochrome P450 2J2: Potential role in drug metabolism and cardiotoxicity. Drug Metab. Dispos. 2018, 46, 1053–1065. [Google Scholar] [CrossRef] [PubMed]
- Campbell, W.B.; Falck, J.R. Arachidonic acid metabolites as endothelium-derived hyperpolarizing factors. Hypertension 2007, 49, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Node, K.; Huo, Y.; Ruan, X.; Yang, B.; Spiecker, M.; Ley, K.; Zeldin, D.C.; Liao, J.K. Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science 1999, 285, 1276–1279. [Google Scholar] [CrossRef] [PubMed]
- Node, K.; Ruan, X.L.; Dai, J.; Yang, S.X.; Graham, L.; Zeldin, D.C.; Liao, J.K. Activation of Gas mediates induction of tissue-type plasminogen activator gene transcription by epoxyeicosatrienoic acids. J. Biol. Chem. 2001, 276, 15983–15989. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Graham, L.; Dikalov, S.; Mason, R.P.; Falck, J.R.; Liao, J.K.; Zeldin, D.C. Overexpression of cytochrome P450 CYP2J2 protects against hypoxia-reoxygenation injury in cultured bovine aortic endothelial cells. Mol. Pharmacol. 2001, 60, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Seubert, J.; Yang, B.; Bradbury, J.A.; Graves, J.; Degraff, L.M.; Gabel, S.; Gooch, R.; Foley, J.; Newman, J.; Mao, L.; et al. Enhanced postischemic functional recovery in CYP2J2 transgenic hearts involves mitochondrial ATP-sensitive K+ channels and p42/p44 MAPK pathway. Circ. Res. 2004, 95, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Spiecker, M.; Liao, J.K. Vascular protective effects of cytochrome p450 epoxygenase-derived eicosanoids. Arch. Biochem. Biophys. 2005, 433, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Theken, K.N.; Lee, C.R. Genetic variation in the cytochrome P450 epoxygenase pathway and cardiovascular disease risk. Pharmacogenomics 2007, 8, 1369–1383. [Google Scholar] [CrossRef] [PubMed]
- Spiecker, M.; Darius, H.; Hankeln, T.; Soufi, M.; Sattler, A.M.; Schaefer, J.R.; Node, K.; Borgel, J.; Mugge, A.; Lindpaintner, K.; et al. Risk of coronary artery disease associated with polymorphism of the cytochrome P450 epoxygenase CYP2J2. Circulation 2004, 110, 2132–2136. [Google Scholar] [CrossRef] [PubMed]
- Ke, Q.; Xiao, Y.F.; Bradbury, J.A.; Graves, J.P.; Degraff, L.M.; Seubert, J.M.; Zeldin, D.C. Electrophysiological properties of cardiomyocytes isolated from CYP2J2 transgenic mice. Mol. Pharmacol. 2007, 72, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Ye, D.; Wang, X.; Seubert, J.M.; Graves, J.P.; Bradbury, J.A.; Zeldin, D.C.; Lee, H.C. Cardiac and vascular KATP channels in rats are activated by endogenous epoxyeicosatrienoic acids through different mechanisms. J. Physiol. 2006, 575, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.F.; Ke, Q.; Seubert, J.M.; Bradbury, J.A.; Graves, J.; Degraff, L.M.; Falck, J.R.; Krausz, K.; Gelboin, H.V.; Morgan, J.P.; et al. Enhancement of cardiac L-type Ca2+ currents in transgenic mice with cardiac-specific overexpression of CYP2J2. Mol. Pharmacol. 2004, 66, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
- Askari, A.; Thomson, S.J.; Edin, M.L.; Zeldin, D.C.; Bishop-Bailey, D. Roles of the epoxygenase CYP2J2 in the endothelium. Prostaglandins Other Lipid Mediat. 2013, 107, 56–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Zhang, X.A.; Wang, D.W. The roles of CYP450 epoxygenases and metabolites, epoxyeicosatrienoic acids, in cardiovascular and malignant diseases. Adv. Drug Deliv. Rev. 2011, 63, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wei, X.; Xiao, X.; Hui, R.; Card, J.W.; Carey, M.A.; Wang, D.W.; Zeldin, D.C. Arachidonic acid epoxygenase metabolites stimulate endothelial cell growth and angiogenesis via mitogen-activated protein kinase and phosphatidylinositol 3-kinase/Akt signaling pathways. J. Pharmacol. Exp. Ther. 2005, 314, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, U.R.; Fleming, I. From endothelium-derived hyperpolarizing factor (EDHF) to angiogenesis: Epoxyeicosatrienoic acids (EETs) and cell signaling. Pharmacol. Ther. 2006, 111, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.G.; Chen, C.L.; Card, J.W.; Yang, S.; Chen, J.X.; Fu, X.N.; Ning, Y.G.; Xiao, X.; Zeldin, D.C.; Wang, D.W. Cytochrome P450 2J2 promotes the neoplastic phenotype of carcinoma cells and is up-regulated in human tumors. Cancer Res. 2005, 65, 4707–4715. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.G.; Ning, Y.G.; Chen, C.; Ma, D.; Liu, Z.J.; Yang, S.; Zhou, J.; Xiao, X.; Zhang, X.A.; Edin, M.L.; et al. Cytochrome p450 epoxygenase promotes human cancer metastasis. Cancer Res. 2007, 67, 6665–6674. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, D.W. CYP epoxygenase derived EETs: From cardiovascular protection to human cancer therapy. Curr. Top. Med. Chem. 2013, 13, 1454–1469. [Google Scholar] [CrossRef] [PubMed]
- Zeldin, D.C. Epoxygenase pathways of arachidonic acid metabolism. J. Biol. Chem. 2001, 276, 36059–36062. [Google Scholar] [CrossRef] [PubMed]
- Westphal, C.; Konkel, A.; Schunck, W.H. CYP-eicosanoids—A new link between omega-3 fatty acids and cardiac disease? Prostaglandins Other Lipid Mediat. 2011, 96, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Arnold, W.R.; Baylon, J.L.; Tajkhorshid, E.; Das, A. Asymmetric Binding and Metabolism of Polyunsaturated Fatty Acids (PUFAs) by CYP2J2 Epoxygenase. Biochemistry 2016, 55, 6969–6980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scarborough, P.E.; Ma, J.; Qu, W.; Zeldin, D.C. P450 subfamily CYP2J and their role in the bioactivation of arachidonic acid in extrahepatic tissues. Drug Metab. Rev. 1999, 31, 205–234. [Google Scholar] [CrossRef] [PubMed]
- McDougle, D.R.; Kambalyal, A.; Meling, D.D.; Das, A. Endocannabinoids anandamide and 2-arachidonoylglycerol are substrates for human CYP2J2 epoxygenase. J. Pharmacol. Exp. Ther. 2014, 351, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Zelasko, S.; Arnold, W.R.; Das, A. Endocannabinoid metabolism by cytochrome P450 monooxygenases. Prostaglandins Other Lipid Mediat. 2015, 116–117, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Arnold, W.R.; Weigle, A.T.; Das, A. Cross-talk of cannabinoid and endocannabinoid metabolism is mediated via human cardiac CYP2J2. J. Inorg. Biochem. 2018, 184, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Walker, V.J.; Griffin, A.P.; Hammar, D.K.; Hollenberg, P.F. Metabolism of Anandamide by Human Cytochrome P450 2J2 in the Reconstituted System and Human Intestinal Microsomes. J. Pharmacol. Exp. Ther. 2016, 357, 537–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.A.; Neul, D.; Clouser-Roche, A.; Dalvie, D.; Wester, M.R.; Jiang, Y.; Jones, J.P., 3rd; Freiwald, S.; Zientek, M.; Totah, R.A. Identification of novel substrates for human cytochrome P450 2J2. Drug Metab. Dispos. 2010, 38, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Hashizume, T.; Mise, M.; Matsumoto, S.; Terauchi, Y.; Fujii, T.; Imaoka, S.; Funae, Y.; Kamataki, T.; Miyazaki, H. A novel cytochrome P450 enzyme responsible for the metabolism of ebastine in monkey small intestine. Drug Metab. Dispos. 2001, 29, 798–805. [Google Scholar] [PubMed]
- Hashizume, T.; Imaoka, S.; Mise, M.; Terauchi, Y.; Fujii, T.; Miyazaki, H.; Kamataki, T.; Funae, Y. Involvement of CYP2J2 and CYP4F12 in the metabolism of ebastine in human intestinal microsomes. J. Pharmacol. Exp. Ther. 2002, 300, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.H.; Kim, M.G.; Lee, D.J.; Yoon, Y.J.; Kim, M.J.; Shon, J.H.; Choi, C.S.; Choi, Y.K.; Desta, Z.; Shin, J.G. Characterization of ebastine, hydroxyebastine, and carebastine metabolism by human liver microsomes and expressed cytochrome P450 enzymes: Major roles for CYP2J2 and CYP3A. Drug Metab. Dispos. 2006, 34, 1793–1797. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Hirama, T.; Kim, H.J.; Nagata, K.; Yamazoe, Y. In vitro inhibition of human small intestinal and liver microsomal astemizole O-demethylation: Different contribution of CYP2J2 in the small intestine and liver. Xenobiotica 2003, 33, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lee, D.; Joo, J.; Shin, J.H.; Kang, W.; Oh, S.; Lee, D.Y.; Lee, S.J.; Yea, S.S.; Lee, H.S.; et al. CYP2J2 and CYP2C19 are the major enzymes responsible for metabolism of albendazole and fenbendazole in human liver microsomes and recombinant P450 assay systems. Antimicrob. Agents Chemother. 2013, 57, 5448–5456. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.A.; Jones, J.P., 3rd; Katayama, J.; Kaspera, R.; Jiang, Y.; Freiwald, S.; Smith, E.; Walker, G.S.; Totah, R.A. Identifying a selective substrate and inhibitor pair for the evaluation of CYP2J2 activity. Drug Metab. Dispos. 2012, 40, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Kaspera, R.; Kirby, B.J.; Sahele, T.; Collier, A.C.; Kharasch, E.D.; Unadkat, J.D.; Totah, R.A. Investigating the contribution of CYP2J2 to ritonavir metabolism in vitro and in vivo. Biochem. Pharmacol. 2014, 91, 109–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lafite, P.; Dijols, S.; Buisson, D.; Macherey, A.C.; Zeldin, D.C.; Dansette, P.M.; Mansuy, D. Design and synthesis of selective, high-affinity inhibitors of human cytochrome P450 2J2. Bioorg. Med. Chem. Lett. 2006, 16, 2777–2780. [Google Scholar] [CrossRef] [PubMed]
- Lafite, P.; Dijols, S.; Zeldin, D.C.; Dansette, P.M.; Mansuy, D. Selective, competitive and mechanism-based inhibitors of human cytochrome P450 2J2. Arch. Biochem. Biophys. 2007, 464, 155–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Li, G.; Liao, W.; Wu, J.; Liu, L.; Ma, D.; Zhou, J.; Elbekai, R.H.; Edin, M.L.; Zeldin, D.C.; et al. Selective inhibitors of CYP2J2 related to terfenadine exhibit strong activity against human cancers in vitro and in vivo. J. Pharmacol. Exp. Ther. 2009, 329, 908–918. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Zeng, J.; Mei, Y.; Zhang, J.Z.; Yan, S.F.; Fei, J.; Chen, L. Discovery and characterization of novel, potent, and selective cytochrome P450 2J2 inhibitors. Drug Metab. Dispos. 2013, 41, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.L.; Zhang, H.; Walker, V.J.; D’Agostino, J.; Hollenberg, P.F. Heme Modification Contributes to the Mechanism-Based Inactivation of Human Cytochrome P450 2J2 by Two Terminal Acetylenic Compounds. Drug Metab. Dispos. 2017, 45, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.L.; Zhang, H.; Hollenberg, P.F. Formation of Both Heme and Apoprotein Adducts Contributes to the Mechanism-Based Inactivation of Human CYP2J2 by 17alpha-Ethynylestradiol. Drug Metab. Dispos. 2018, 46, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Lafite, P.; Andre, F.; Zeldin, D.C.; Dansette, P.M.; Mansuy, D. Unusual regioselectivity and active site topology of human cytochrome P450 2J2. Biochemistry 2007, 46, 10237–10247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Tang, Y.; Liu, H.; Cheng, J.; Zhu, W.; Jiang, H. Probing ligand binding modes of human cytochrome P450 2J2 by homology modeling, molecular dynamics simulation, and flexible molecular docking. Proteins 2008, 71, 938–949. [Google Scholar] [CrossRef] [PubMed]
- Cong, S.; Ma, X.T.; Li, Y.X.; Wang, J.F. Structural basis for the mutation-induced dysfunction of human CYP2J2: A computational study. J. Chem. Inf. Model. 2013, 53, 1350–1357. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.L.; Fa, B.T.; Cong, S.; Wang, J.F.; Chou, K.C. Research/review: Insights into the mutation-induced dysfunction of arachidonic acid metabolism from modeling of human CYP2J2. Curr. Drug Metab. 2014, 15, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Proietti, G.; Abelak, K.K.; Bishop-Bailey, D.; Macchiarulo, A.; Nobeli, I. Computational modelling of the binding of arachidonic acid to the human monooxygenase CYP2J2. J. Mol. Model. 2016, 22, 279. [Google Scholar] [CrossRef] [PubMed]
- Arnold, W.R.; Baylon, J.L.; Tajkhorshid, E.; Das, A. Arachidonic Acid Metabolism by Human Cardiovascular CYP2J2 Is Modulated by Doxorubicin. Biochemistry 2017, 56, 6700–6712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, L.M.; Ma, J.; Srettabunjong, S.; Graves, J.; Bradbury, J.A.; Li, L.; Spiecker, M.; Liao, J.K.; Mohrenweiser, H.; Zeldin, D.C. Cloning of CYP2J2 gene and identification of functional polymorphisms. Mol. Pharmacol. 2002, 61, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Chen, W.; Murphy, E.; Gabel, S.; Tomer, K.B.; Foley, J.; Steenbergen, C.; Falck, J.R.; Moomaw, C.R.; Zeldin, D.C. Molecular cloning, expression, and functional significance of a cytochrome P450 highly expressed in rat heart myocytes. J. Biol. Chem. 1997, 272, 12551–12559. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Lindberg, R.L.; Juvonen, R.O.; Negishi, M. Site-directed mutagenesis of mouse steroid 7 alpha-hydroxylase (cytochrome P-450(7) alpha): Role of residue-209 in determining steroid-cytochrome P-450 interaction. Biochem. J. 1993, 291, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Imaoka, S.; Ogawa, H.; Kimura, S.; Gonzalez, F.J. Complete cDNA sequence and cDNA-directed expression of CYP4A11, a fatty acid omega-hydroxylase expressed in human kidney. DNA Cell Biol. 1993, 12, 893–899. [Google Scholar] [PubMed]
- Cheng, Y.; Prusoff, W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar] [PubMed]
- Johnson, E.F.; Stout, C.D. Structural diversity of human xenobiotic-metabolizing cytochrome P450 monooxygenases. Biochem. Biophys. Res. Commun. 2005, 338, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Poulos, T.L.; Johnson, E.F. Structures of cytochromes P450 enzymes. In Cytochrome P450: Structure, Mechanism, and Biochemistry, 3rd ed.; Ortiz de Montellano, P.R., Ed.; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2005; pp. 87–114. [Google Scholar]
- Ma, J.; Qu, W.; Scarborough, P.E.; Tomer, K.B.; Moomaw, C.R.; Maronpot, R.; Davis, L.S.; Breyer, M.D.; Zeldin, D.C. Molecular cloning, enzymatic characterization, developmental expression, and cellular localization of a mouse cytochrome P450 highly expressed in kidney. J. Biol. Chem. 1999, 274, 17777–17788. [Google Scholar] [CrossRef] [PubMed]
- Omura, T.; Sato, R. The Carbon Monoxide-Binding Pigment of Liver Microsomes. I. Evidence for Its Hemoprotein Nature. J. Biol. Chem. 1964, 239, 2370–2378. [Google Scholar] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, C. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Gould, I.R.; Merz, K.M.; Ferguson, D.M.; Spellmeyer, D.C.; Fox, T.; Caldwell, J.W.; Kollman, P.A. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995, 117, 5179–5197. [Google Scholar] [CrossRef] [Green Version]
- Oda, A.; Yamaotsu, N.; Hirono, S. New AMBER force field parameters of heme iron for cytochrome P450s determined by quantum chemical calculations of simplified models. J. Comput. Chem. 2005, 26, 818–826. [Google Scholar] [PubMed]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Delaforge, M.; Loiseau, N. A Method for Performing Restrained Dynamics Docking of One or Multiple Substrates on Multi-Specific Enzymes. Patent No. WO2004038655, 6 May 2004. [Google Scholar]




| Kinetic Constants | WT | R117K | R117L | R117E |
|---|---|---|---|---|
| KM (µM) | 0.3 ± 0.1 | 0.4 ± 0.1 | 1.0 ± 0.2 | 0.1 ± 0.05 |
| kcat (min−1) | 40 ± 5 | 102 ± 3 | 12 ± 2 | 27 ± 3 |
| kcat/KM (min−1 µM−1) | 133 | 245 | 12 | 193 |

| Compound | Y- | -X- | -R | IC50 (µM) a | |||
|---|---|---|---|---|---|---|---|
| WT b | R117K | R117L | R117E | ||||
| 3 | |||||||
| Terfenadone 1 | Ph2(OH)C- | -CO- | --C(CH3)3 | 0.7 ± 0.1 | 1.6 ± 0.1 | 1.9 ± 0.2 | 1.0 ± 0.3 |
| 2 | Ph2(OH)C- | -CO- | (-(CH2)2-CH3 | 0.4 ± 0.1 | 1.1 ± 0.2 | 2.4 ± 0.3 | 0.5 ± 0.3 |
| 3 | Ph2(OH)C- | -CH2- | --C(CH3)3 | 3.6 ± 0.7 | 8.6 ± 0.2 | 2.2 ± 0.2 | 2.1 ± 0.4 |
| 4 | Ph2(OH)C- | -CH2- | --(CH2)2-CH3 | 4.5 ± 0.9 | 13.2 ± 0.1 | 2.3 ± 0.3 | 2.9 ± 0.2 |

| Substrate | Regioselectivity (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | R117K | R117L | R117E | |||||||||||
| -X- | -R | α | β | γ | α | β | γ | α | β | γ | α | β | γ | |
| 2 | -CO- |  | 4 | 87 | 9 | 4 | 92 | 4 | 15 | 79 | 6 | 5 | 90 | 5 |
| 4 | -CH2- |  | 35 | 64 | <1 | 33 | 64 | 2 | 44 | 52 | 4 | 13 | 83 | 1 |
| 5 | -CO- |  | 3 | 97 | 4 | 96 | 17 | 83 | 3 | 97 | ||||
| Substrate | Fe-C Distances (Å)a | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT b | R117K | R117L | R117E | ||||||||||
| -X- | -R | α | β | γ | α | β | γ | α | β | γ | α | β | γ |
| -CO- |  | 4.9 | 3.8 | 4.6 | 5.3 | 4.0 | 4.4 | 3.9 | 3.6 | 3.6 | 5.4 | 4.2 | 4.8 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lafite, P.; André, F.; Graves, J.P.; Zeldin, D.C.; Dansette, P.M.; Mansuy, D. Role of Arginine 117 in Substrate Recognition by Human Cytochrome P450 2J2. Int. J. Mol. Sci. 2018, 19, 2066. https://doi.org/10.3390/ijms19072066
Lafite P, André F, Graves JP, Zeldin DC, Dansette PM, Mansuy D. Role of Arginine 117 in Substrate Recognition by Human Cytochrome P450 2J2. International Journal of Molecular Sciences. 2018; 19(7):2066. https://doi.org/10.3390/ijms19072066
Chicago/Turabian StyleLafite, Pierre, François André, Joan P. Graves, Darryl C. Zeldin, Patrick M. Dansette, and Daniel Mansuy. 2018. "Role of Arginine 117 in Substrate Recognition by Human Cytochrome P450 2J2" International Journal of Molecular Sciences 19, no. 7: 2066. https://doi.org/10.3390/ijms19072066
APA StyleLafite, P., André, F., Graves, J. P., Zeldin, D. C., Dansette, P. M., & Mansuy, D. (2018). Role of Arginine 117 in Substrate Recognition by Human Cytochrome P450 2J2. International Journal of Molecular Sciences, 19(7), 2066. https://doi.org/10.3390/ijms19072066