Changes in the Proteome of Medicago sativa Leaves in Response to Long-Term Cadmium Exposure Using a Cell-Wall Targeted Approach
Abstract
:1. Introduction
2. Results
3. Discussion
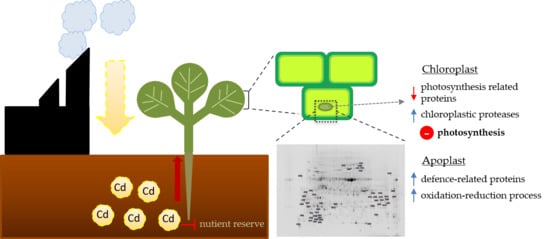
3.1. Cd-Induced Degradation of Photosynthetic Proteins
3.2. Cd Influences the Abundance of Proteins Related to the Cell Wall Structure
3.3. Enhanced Accumulation of Defence Proteins as a Response to Cd
4. Materials and Methods
4.1. Plant Material and Treatment
4.2. Cell Wall Protein Enrichment
4.3. Quantitative Protein Analysis and Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 2D DIGE | Two-dimensional difference gel electrophoresis |
| Ca | Calcium |
| Cd | Cadmium |
| DW | Dry weight |
| HG | Homogalacturonan |
| kDa | Kilo Dalton |
| MALDI | Matrix-assisted Laser Desorption/Ionization |
| MS | Mass spectrum |
| MW | Molecular weight |
| PCA | Principal component analysis |
| PME | Pectin methylesterase |
| ROS | Reactive oxygen species |
| RuBisCO | Ribulose bisphosphate carboxylase/oxidase |
| SAR | Systemic acquired resistance |
| TOF | Time of flight |
References
- Kovalchuk, O.; Titov, V.; Hohn, B.; Kovalchuk, I. A sensitive transgenic plant system to detect toxic inorganic compounds in the environment. Nat. Biotechnol. 2001, 19, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Di Toppi, L.S.; Gabbrielli, R. Response to cadmium in higher plants. Environ. Exp. Bot. 1999, 41, 105–130. [Google Scholar] [CrossRef]
- Chugh, L.K.; Sawhney, S.K. Photosynthetic activities of Pisum sativum seedlings grown in presence of cadmium. Plant Physiol. Biochem. 1999, 37, 297–303. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, X.; Li, K.; Wu, M.; Zhang, R.; Zhang, L.; Chen, G. Photosynthetic responses of Oryza sativa L. seedlings to cadmium stress: Physiological, biochemical and ultrastructural analyses. Biometals 2014, 27, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Faller, P.; Kienzler, K.; Krieger-Liszkay, A. Mechanism of Cd2+ toxicity: Cd2+ inhibits photoactivation of Photosystem II by competitive binding to the essential Ca2+ site. Biochim. Biophys. Acta Bioenerg. 2005, 1706, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Cassab, G.I. Plant Cell Wall Proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 281–309. [Google Scholar] [CrossRef] [PubMed]
- Fry, S.C.; Smith, R.C.; Renwick, K.F.; Martin, D.J.; Hodge, S.K.; Matthews, K.J. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 1992, 282, 821–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Q.; Bergmann, C.W.; Rose, J.K.C.; Saladie, M.; Kolli, V.S.K.; Albersheim, P.; Darvill, A.G.; York, W.S. Characterization of a tomato protein that inhibits a xyloglucan-specific endoglucanase. Plant J. 2003, 34, 327–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradley, D.J.; Kjellbom, P.; Lamb, C.J. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: A novel, rapid defense response. Cell 1992, 70, 21–30. [Google Scholar] [CrossRef]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef] [PubMed]
- Krzesłowska, M. The cell wall in plant cell response to trace metals: Polysaccharide remodeling and its role in defense strategy. Acta Physiol. Plant 2011, 33, 35–51. [Google Scholar] [CrossRef]
- Vollenweider, P.; Cosio, C.; Günthardt-Goerg, M.S.; Keller, C. Localization and effects of cadmium in leaves of a cadmium-tolerant willow (Salix viminalis L.). Environ. Exp. Bot. 2006, 58, 25–40. [Google Scholar] [CrossRef]
- Parrotta, L.; Guerriero, G.; Sergeant, K.; Cai, G.; Hausman, J.-F. Target or barrier? The cell wall of early- and later-diverging plants vs. cadmium toxicity: Differences in the response mechanisms. Front. Plant Sci. 2015, 6, 133. [Google Scholar] [CrossRef] [PubMed]
- Douchiche, O.; Soret-Morvan, O.; Chaïbi, W.; Morvan, C.; Paynel, F. Characteristics of cadmium tolerance in “Hermes” flax seedlings: Contribution of cell walls. Chemosphere 2010, 81, 1430–1436. [Google Scholar] [CrossRef] [PubMed]
- Chaoui, A.; El Ferjani, E. Effects of cadmium and copper on antioxidant capacities, lignification and auxin degradation in leaves of pea (Pisum sativum L.) seedlings. C. R. Biol. 2005, 328, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Paynel, F.; Schaumann, A.; Arkoun, M.; Douchiche, O.; Morvan, C. Temporal regulation of cell-wall pectin methylesterase and peroxidase isoforms in cadmium-treated flax hypocotyl. Ann. Bot. 2009, 104, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Albenne, C.; Canut, H.; Jamet, E. Plant cell wall proteomics: The leadership of Arabidopsis thaliana. Front. Plant Sci. 2013, 4, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Haslam, R.P.; Downie, A.L.; Raveton, M.; Gallardo, K.; Job, D.; Pallett, K.E.; John, P.; Parry, M.A.J.; Coleman, J.O.D. The assessment of enriched apoplastic extracts using proteomic approaches. Ann. Appl. Biol. 2003, 143, 81–91. [Google Scholar] [CrossRef]
- Ndimba, B.K.; Chivasa, S.; Hamilton, J.M.; Simon, W.J.; Slabas, A.R. Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics 2003, 3, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Plaxton, W.C. Proteomic analysis of alterations in the secretome of Arabidopsis thaliana suspension cells subjected to nutritional phosphate deficiency. Proteomics 2008, 8, 4317–4326. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Song, T.; Fan, H.; Yu, Y.; Cui, N.; Zhao, J.; Meng, K. A comparative cell wall proteomic analysis of cucumber leaves under Sphaerotheca fuliginea stress. Acta Physiol. Plant. 2016, 38, 260. [Google Scholar] [CrossRef]
- Dani, V.; Simon, W.J.; Duranti, M.; Croy, R.R.D. Changes in the tobacco leaf apoplast proteome in response to salt stress. Proteomics 2005, 5, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Printz, B.; Guerriero, G.; Sergeant, K.; Audinot, J.-N.; Guignard, C.; Renaut, J.; Lutts, S.; Hausman, J.-F. Combining-Omics to Unravel the Impact of Copper Nutrition on Alfalfa (Medicago sativa) Stem Metabolism. Plant Cell Physiol. 2016, 57, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, J.C.; Hatfield, R.D.; Sullivan, M.L. Proteomic analysis of cell walls of two developmental stages of alfalfa stems. Front. Plant Sci. 2012, 3, 279. [Google Scholar] [CrossRef] [PubMed]
- Sergeant, K.; Printz, B.; Gutsch, A.; Behr, M.; Renaut, J.; Hausman, J.-F. Didehydrophenylalanine, an abundant modification in the β subunit of plant polygalacturonases. PLoS ONE 2017, 12, e0171990. [Google Scholar] [CrossRef] [PubMed]
- Printz, B.; Morais, R.D.S.; Wienkoop, S.; Sergeant, K.; Lutts, S.; Hausman, J.-F.; Renaut, J. An improved protocol to study the plant cell wall proteome. Front. Plant Sci. 2015, 6, 237. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Sun, Y.; Yang, Q.; Tian, H.; Zhang, H.; Liu, Y.; Chen, M. Proteomic analysis of early salt stress responsive proteins in alfalfa roots and shoots. Proteome Sci. 2017, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, J.-M.; Pietrzak, M.; Boller, T. Mutation analysis of the C-terminal vacuolar targeting peptide of tobacco chitinase: Low specificity of the sorting system, and gradual transition between intracellular retention and secretion into the extracellular space. Plant J. 1994, 5, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Stigliano, E.; Di Sansebastiano, G.P.; Neuhaus, J.M. Contribution of chitinase A’s C-terminal vacuolar sorting determinant to the study of soluble protein compartmentation. Int. J. Mol. Sci. 2014, 15, 11030–11039. [Google Scholar] [CrossRef] [PubMed]
- Gutsch, A.; Keunen, E.; Guerriero, G.; Renaut, J.; Cuypers, A.; Hausman, J.-F.; Sergeant, K. Long-term cadmium exposure influences the abundance of proteins that impact the cell wall structure in Medicago sativa stems. Plant Biol. J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Videa, J.R.; De la Rosa, G.; Gonzalez, J.H.; Gardea-Torresdey, J.L. Effects of the growth stage on the heavy metal tolerance of alfalfa plants. Adv. Environ. Res. 2004, 8, 679–685. [Google Scholar] [CrossRef]
- Wang, C.Q.; Song, H. Calcium protects Trifolium repens L. seedlings against cadmium stress. Plant Cell Rep. 2009, 28, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Gratão, P.L.; Monteiro, C.C.; Antunes, A.M.; Peres, L.E.P.; Azevedo, R.A. Acquired tolerance of tomato (Lycopersicon esculentum cv. Micro-Tom) plants to cadmium-induced stress. Ann. Appl. Biol. 2008, 153, 321–333. [Google Scholar] [CrossRef]
- Schützendübel, A.; Polle, A. Plant responses to abiotic stresses: Heavy metal-induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 2002, 53, 1351–1365. [Google Scholar] [CrossRef] [PubMed]
- Romero-Puertas, M.C.; Palma, J.M.; Gómez, M.; Río, L.A.D.E.L.; Sandalio, L.M. Cadmium causes the oxidative modification of proteins in pea plants. Plant Cell Environ. 2002, 25, 677–686. [Google Scholar] [CrossRef] [Green Version]
- Sandalio, L.M.; Dalurzo, H.C.; Gómez, M.; Romero-Puertas, M.C.; del Río, L.A. Cadmium-induced changes in the growth and oxidative metabolism of pea plants. J. Exp. Bot. 2001, 52, 2115–2126. [Google Scholar] [CrossRef] [PubMed]
- Semane, B.; Dupae, J.; Cuypers, A.; Noben, J.P.; Tuomainen, M.; Tervahauta, A.; Kärenlampi, S.; van Belleghem, F.; Smeets, K.; Vangronsveld, J. Leaf proteome responses of Arabidopsis thaliana exposed to mild cadmium stress. J. Plant Physiol. 2010, 167, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Simões, I.; Faro, C. Structure and function of plant aspartic proteinases. Eur. J. Biochem. 2004, 271, 2067–2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhalerao, R.; Keskitalo, J.; Sterky, F.; Erlandsson, R.; Björkbacka, H.; Birve, S.J.; Karlsson, J.; Gardeström, P.; Gustafsson, P.; Lundeberg, J.; et al. Gene expression in autumn leaves. Plant Physiol. 2003, 131, 430–442. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Murakami, S.; Yamamoto, Y.; Chatani, H.; Kondo, Y.; Nakano, T.; Yokota, A.; Sato, F. The DNA-binding protease, CND41, and the degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase in senescent leaves of tobacco. Planta 2004, 220, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Parrott, D.L.; McInnerney, K.; Feller, U.; Fischer, A.M. Steam-girdling of barley (Hordeum vulgare) leaves leads to carbohydrate accumulation and accelerated leaf senescence, facilitating transcriptomic analysis of senescence-associated genes. New Phytol. 2007, 176, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Radlowski, M.; Kalinowski, A.; Adamczyk, J.; Krolikowski, Z.; Bartkowiak, S. Proteolytic activity in the maize pollen wall. Physiol. Plant. 1996, 98, 172–178. [Google Scholar] [CrossRef]
- Xia, Y.; Suzuki, H.; Borevitz, J.; Blount, J.; Guo, Z.; Patel, K.; Dixon, R.A.; Lamb, C. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 2004, 23, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Breitenbach, H.H.; Wenig, M.; Wittek, F.; Jorda, L.; Maldonado-Alconada, A.M.; Sarioglu, H.; Colby, T.; Knappe, C.; Bichlmeier, M.; Pabst, E.; et al. Contrasting roles of the apoplastic aspartyl protease apoplastic, enhanced disease susceptibility1-dependent1 and legume lectin-like protein1 in Arabidopsis systemic acquired resistance. Plant Physiol. 2014, 165, 791–809. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ding, Y.; Wang, F.; Ye, Y.; Zhu, C. Role of salicylic acid in resistance to cadmium stress in plants. Plant Cell Rep. 2016, 35, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Micheli, F. Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 2001, 6, 414–419. [Google Scholar] [CrossRef]
- Ramos, I.; Esteban, E.; Lucena, J.J.; Gárate, A. Cadmium uptake and subcellular distribution in plants of Lactuca sp. Cd-Mn interaction. Plant Sci. 2002, 162, 761–767. [Google Scholar] [CrossRef]
- Douchiche, O.; Driouich, A.; Morvan, C. Spatial regulation of cell-wall structure in response to heavy metal stress: Cadmium-induced alteration of the methyl-esterification pattern of homogalacturonans. Ann. Bot. 2010, 105, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Mouille, G.; Pelloux, J. Homogalacturonan methyl-esterification and plant development. Mol. Plant 2009, 2, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H.; Ma, Y.; Chen, N.; Guo, S.; Liu, H.H.; Guo, X.; Chong, K.; Xu, Y. Overexpression of stress-inducible OsBURP16, the β subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice. Plant Cell Environ. 2014, 37, 1144–1158. [Google Scholar] [CrossRef] [PubMed]
- Watson, C.F.; Zheng, L.; DellaPenna, D. Reduction of tomato polygalacturonase β subunit expression affects pectin solubilization and degradation during fruit ripening. Plant Cell 1994, 6, 1623–1634. [Google Scholar] [CrossRef] [PubMed]
- Tucker, G.A.; Robertson, N.G.; Grierson, D. The conversion of tomato-fruit polygalacturonase isoenzyme 2 into isoenzyme 1 in vitro. Eur. J. Biochem. 1981, 115, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Favaron, F.; D’Ovidio, R.; Alghisi, P. Purification and molecular charecterisation of a soybean polygalacturonase-inhibiting protein. Planta 1994, 195, 80–87. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzo, G.; Ferrari, S. Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr. Opin. Plant Biol. 2002, 5, 295–299. [Google Scholar] [CrossRef]
- Spadoni, S.; Zabotina, O.; di Matteo, A.; Mikkelsen, J.D.; Cervone, F.; de Lorenzo, G.; Mattei, B.; Bellincampi, D. Polygalacturonase-inhibiting protein interacts with pectin through a binding site formed by four clustered residues of arginine and lysine. Plant Physiol. 2006, 141, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, P.; Schröder, P.; Dommes, J.; Hoffmann, L.; Renaut, J.; Hausman, J.F. Proteomic and enzymatic response of poplar to cadmium stress. J. Proteom. 2009, 72, 379–396. [Google Scholar] [CrossRef]
- Loix, C.; Huybrechts, M.; Vangronsveld, J.; Gielen, M.; Keunen, E.; Cuypers, A. Reciprocal interactions between cadmium-induced cell wall responses and oxidative stress in plants. Front. Plant Sci. 2017, 8, 1867. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef] [PubMed]
- McDougall, G.J. Changes in cell wall-associated peroxidases during the lignification of flax fibres. Phytochemistry 1992, 31, 3385–3389. [Google Scholar] [CrossRef]
- Chaoui, A.; Jarrar, B.; El Ferjani, E. Effects of cadmium and copper on peroxidase, NADH oxidase and IAA oxidase activities in cell wall, soluble and microsomal membrane fractions of pea roots. J. Plant Physiol. 2004, 161, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Radotic, K.; Ducic, T.; Mutavdzic, D. Changes in peroxidase activity and isoenzymes in spruce needles after exposure to different concentrations of cadmium. Environ. Exp. Bot. 2000, 44, 105–113. [Google Scholar] [CrossRef]
- Luhua, S.; Ciftci-Yilmaz, S.; Harper, J.; Cushman, J.; Mittler, R. Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function. Plant Physiol. 2008, 148, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Kamiya, T.; Kalmbach, L.; Yamagami, M.; Yamaguchi, K.; Shigenobu, S.; Sawa, S.; Danku, J.M.C.; Salt, D.E.; Geldner, N.; et al. Role of LOTR1 in nutrient transport through organization of spatial distribution of root endodermal barriers. Curr. Biol. 2017, 27, 758–765. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yan, W.; Sun, L.; Tian, J.; Liao, H. Proteomic analysis reveals growth inhibition of soybean roots by manganese toxicity is associated with alteration of cell wall structure and lignification. J. Proteomics 2016, 143, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Békésiová, B.; Hraška, Š.; Libantová, J.; Moravčíková, J.; Matušíková, I. Heavy-metal stress induced accumulation of chitinase isoforms in plants. Mol. Biol. Rep. 2008, 35, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Bernier, F.; Berna, A. Germins and germin-like proteins: Plant do-all proteins. But what do they do exactly? Plant Physiol. Biochem. 2001, 39, 545–554. [Google Scholar] [CrossRef]
- The Alfalfa Gene Index and Expression Atlas Dadabase. Available online: http://plantgrn.noble.org/AGED/SearchGene.jsp (accessed on 1 January 2015).
- Emanuelsson, O.; Nielsen, H. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Duruflé, H.; Clemente, H.; Balliau, T.; Zivy, M.; Dunand, C.; Jamet, E. Cell wall proteome analysis of Arabidopsis thaliana mature stems. Proteomics 2017, 17, 1600449. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.; Sønderby, C.; Sønderby, S.; Nielsen, H.; Winther, O. Deep Loc: prediction of protein subcellular localization using deep learning. Bioinformatics 2017, 33, 3387–3395. [Google Scholar] [CrossRef] [PubMed]



| Protein Identification | NCBI Identification * | Nr. of Spots Wherein the Protein Was Identified | TargetP |
|---|---|---|---|
| Lower Abundant in Cd-Exposed Plants | |||
| Carbohydrate Metabolic Process | |||
| Sedoheptulose-1,7-bisphosphatase | gi|357461143 | 1 | C |
| Cell wall modification | |||
| Pectinesterase/pectinesterase inhibitor | gi|357504799 | 3 | S |
| Polygalacturonase non-catalytic protein | gi|922335979 | 10 | S |
| Polygalacturonase-inhibiting protein 1 | gi|374634428 | 1 | / |
| Defence | |||
| Cystatin | gi|74058377 | 1 | / |
| Nod factor-binding lectin-nucleotide phosphohydrolase | gi|357508587 | 4 | S |
| Pathogenesis-related thaumatin family protein | gi|922367846 | 1 | S |
| CAP, cysteine-rich secretory protein, antigen 5 | gi|357446161 | 1 | S |
| Nutrient reserve | |||
| Auxin-binding protein ABP19a | gi|357513969 | 11 | S |
| Germin-like protein subfamily 3 member 1 | gi|502156424 | 1 | S |
| Stem 28 kDa glycoprotein | gi|357513539 | 12 | S |
| Oxidation-reduction process | |||
| 1-cys peroxiredoxin PER1 | gi|922395795 | 1 | C |
| Photosynthesis | |||
| Chlorophyll a-b binding protein 2 | gi|3293555 | 1 | C |
| Oxygen-evolving enhancer protein | gi|922331371 | 6 | C |
| Ribose-5-phosphate isomerase A | gi|357512271 | 4 | C |
| Ribulose bisphosphate carboxylase small chain | gi|3914601 | 5 | C |
| Photosystem I reaction centre subunit IV A | gi|922402507 | 1 | C |
| Photosystem II oxygen-evolving enhancer protein | gi|922336891 | 1 | C |
| Proteolysis | |||
| Eukaryotic aspartyl protease family protein | gi|922379288 | 6 | S |
| Secondary metabolites | |||
| Lactoylglutathione lyase-like protein | gi|922388614 | 1 | / |
| Unknown | |||
| Plant/F18G18-200 protein | gi|922395263 | 6 | S |
| Higher Abundant in Cd-Exposed Plants | |||
| Carbohydrate metabolic process | |||
| Glucan endo-1,3-β-glucosidase | gi|357474061 | 11 | S |
| Glycoside hydrolase, family 17 | gi|87240471 | 1 | / |
| Glycoside hydrolase family 18 protein | gi|357454031 | 3 | S |
| Defence | |||
| Allergen Pru protein, putative | gi|922401927 | 7 | S |
| Chitinase (Class Ib)/Hevein | gi|922329699 | 6 | S |
| Chitinase/Hevein/PR-4/Wheatwin2 | gi|922347233 | 10 | S |
| Chitinase | gi|357443753 | 4 | S |
| Class I chitinase | gi|1800141 | 5 | S |
| Disease resistance response protein | gi|922325015 | 2 | S or/ |
| Pathogenesis-related protein 1 | gi|548592 | 1 | S |
| Pathogenesis-related thaumatin family protein | gi|922338021 | 4 | S |
| Plant basic secretory protein (BSP) family protein | gi|922407517 | 2 | S |
| Pre-hevein-like protein | gi|7381205 | 1 | / |
| Stromal 70 kDa heat shock-related protein | gi|821595433 | 1 | C |
| CAP, cysteine-rich secretory protein, antigen 5 | gi|357446161 | 7 | S |
| Nutrient reserve | |||
| Auxin-binding protein ABP19a | gi|357513969 | 1 | S |
| Rhicadhesin receptor | gi|357511665 | 2 | S |
| Oxidation-reduction process | |||
| Anionic peroxidase swpb3 protein | gi|922380311 | 1 | S |
| Class III peroxidase | gi|357476371 | 10 | S |
| Peroxidase | gi|537317 | 7 | S |
| Peroxidase family protein | gi|357448431 | 1 | S |
| Peroxidase1b | gi|971560 | 3 | S |
| Peroxidase2 | gi|13992528 | 10 | S |
| Plastocyanin-like domain protein | gi|922335020 | 2 | S |
| Photosynthesis | |||
| Photosystem I reaction centre subunit II | gi|357480841 | 1 | C |
| Proteolysis | |||
| Carboxyl-terminal peptidase | gi|922336331 | 2 | S |
| Eukaryotic aspartyl protease family protein | gi|922327497 | 14 | C |
| Papain family cysteine protease | gi|357437715 | 2 | S |
| Polyubiquitin | gi|695063425 | 6 | / |
| Subtilisin-like serine protease | gi|922333118 | 1 | S |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutsch, A.; Zouaghi, S.; Renaut, J.; Cuypers, A.; Hausman, J.-F.; Sergeant, K. Changes in the Proteome of Medicago sativa Leaves in Response to Long-Term Cadmium Exposure Using a Cell-Wall Targeted Approach. Int. J. Mol. Sci. 2018, 19, 2498. https://doi.org/10.3390/ijms19092498
Gutsch A, Zouaghi S, Renaut J, Cuypers A, Hausman J-F, Sergeant K. Changes in the Proteome of Medicago sativa Leaves in Response to Long-Term Cadmium Exposure Using a Cell-Wall Targeted Approach. International Journal of Molecular Sciences. 2018; 19(9):2498. https://doi.org/10.3390/ijms19092498
Chicago/Turabian StyleGutsch, Annelie, Salha Zouaghi, Jenny Renaut, Ann Cuypers, Jean-Francois Hausman, and Kjell Sergeant. 2018. "Changes in the Proteome of Medicago sativa Leaves in Response to Long-Term Cadmium Exposure Using a Cell-Wall Targeted Approach" International Journal of Molecular Sciences 19, no. 9: 2498. https://doi.org/10.3390/ijms19092498
APA StyleGutsch, A., Zouaghi, S., Renaut, J., Cuypers, A., Hausman, J. -F., & Sergeant, K. (2018). Changes in the Proteome of Medicago sativa Leaves in Response to Long-Term Cadmium Exposure Using a Cell-Wall Targeted Approach. International Journal of Molecular Sciences, 19(9), 2498. https://doi.org/10.3390/ijms19092498