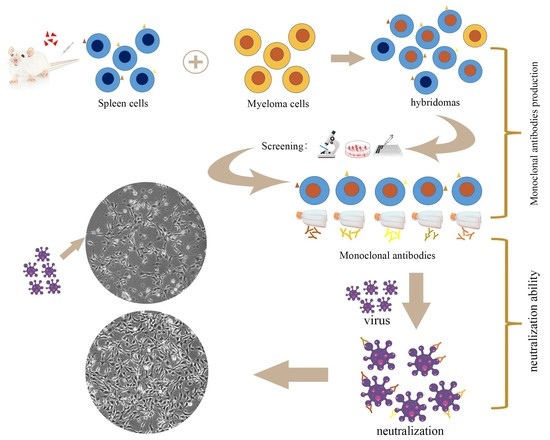
Development and Characterization of Monoclonal Antibodies to the 32 kDa Viral Attachment Protein of Lymphocystis Disease Virus and Their Neutralizing Ability in Vitro
Abstract
:1. Introduction
2. Results
2.1. Expression, Purification and SDS-PAGE Analysis of Recombinant Viral Attachment Protein
2.2. Production, Screening and Subtype Identification of Anti-32 kDa Viral Attachment Protein Monoclonal Antibodies
2.3. Reaction Abilities of Anti-32 kDa Viral Attachment Protein Monoclonal Antibodies
2.4. Lymphocystis Disease Virus Detection in Flounder (P. Olivaceus) Gill Cells by Anti-32 kDa Viral Attachment Protein Monoclonal Antibodies
2.5. Neutralization Ability of the Monoclonal Antibodies to Lymphocystis Disease Virus Infection
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Cells, Virus and Proteins
4.3. Mouse Immunization and Monoclonal Antibody Production
4.4. Indirect Enzyme-Linked Immunosorbent Assay
4.5. Subtype Identification of the MAbs
4.6. SDS-PAGE and Western Blotting
4.7. Indirect Immunofluorescence Assay
4.8. Preparation of Mice Ascites
4.9. Neutralization Assay
4.10. Statistics
Author Contributions
Funding
Conflicts of Interest
References
- Hossain, M.; Kim, S.R.; Itamura, S.I.; Kim, D.W.; Jung, S.J.; Nishizawa, T.; Yoshimizu, M.; Oh, M.J. Lymphocystis disease virus persists in the epidermal tissues of olive flounder, Paralichthys olivaceus (Temminch & Schlegel), at low temperatures. J. Fish Dis. 2009, 32, 699–703. [Google Scholar] [PubMed]
- Wolf, K. Lymphocystis disease. In Fish Viruses and Fish Viral Diseases; Cornell University Press: Ithaca, NY, USA, 1988; pp. 268–291. [Google Scholar]
- Iwamoto, R.; Hasegawa, O.; LaPatra, S.; Yoshimizu, M. Isolation and characterization of the Japanese flounder (Paralichthys olivaceus) lymphocystis disease virus. J. Aquat. Anim. Health 2002, 14, 114–123. [Google Scholar] [CrossRef]
- Flügel, R.M. Lymphocystis disease virus. Curr. Top. Microbiol. Immunol. 1985, 166, 133–150. [Google Scholar]
- Walker, R. Fine structure of lymphocystis virus of fish. Virology 1962, 18, 503–505. [Google Scholar] [CrossRef]
- Zwillenberg, L.O.; Wolf, K. Ultrastructure of lymphocystis virus. J. Virol. 1968, 2, 393–399. [Google Scholar] [PubMed]
- Schnitzler, P.; Darai, G. Identification of the gene encoding the major capsid protein of fish lymphocystis disease virus. J. Gen. Virol. 1993, 74, 2143–2150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, X.Y.; Wu, Z.H.; Jian, J.C.; Lu, Y.S.; Sun, X.Q. Analysis of the genetic diversity of the lymphocystis virus and its evolutionary relationship with its hosts. Virus Genes 2011, 43, 358. [Google Scholar] [CrossRef] [PubMed]
- Cano, I.; Alonso, M.C.; Garcia-Rosado, E.; Saint-Jean, S.R.; Castro, D.; Borrego, J.J. Detection of lymphocystis disease virus (LCDV) in asymptomatic cultured gilt-head seabream (Sparus aurata, L.) using an immunoblot technique. Vet. Microbiol. 2006, 113, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Iwakiri, S.; Song, J.Y.; Nakayama, K.; Oh, M.J.; Ishida, M.; Kitamura, S. Host responses of Japanese flounder Paralichthys olivaceus with lymphocystis cell formation. Fish Shellfish Immunol. 2014, 38, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Tidona, C.A.; Darai, G. The complete DNA sequence of lymphocystis disease virus. J. Virol. 1997, 238, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Y.; Xiao, F.; Xie, J.; Li, Z.Q.; Gui, J.F. Complete genome sequence of lymphocystis disease virus isolate from China. J. Virol. 2004, 78, 6982–6994. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sheng, X.Z.; Xing, J.; Tang, X.Q.; Zhan, W.B. Identification of a 27.8 kDa protein from flounder gill cells involved in lymphocystis disease virus binding and infection. Dis. Aquat. Organ. 2011, 94, 9–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, X.Z.; Wang, M.; Xing, J.; Tang, X.Q.; Zhan, W.B. Monoclonal antibodies against 27.8 kDa protein receptor efficiently block lymphocystis disease virus infection in flounder Paralichthys olivaceus gill cells. Dis. Aquat. Organ. 2012, 100, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.Z.; Wu, R.H.; Tang, X.Q.; Xing, J.; Zhan, W.B. Tissue localization of lymphocystis disease virus (LCDV) receptor-27.8 kDa and its expression kinetics induced by the viral infection in turbot (Scophthalmus maximus). Int. J. Mol. Sci. 2015, 16, 26506–26519. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Fei, C.J.; Tang, X.Q.; Sheng, X.Z.; Zhan, W.B. A 32 kDa viral attachment protein of lymphocystis disease virus (LCDV) specifically interacts with a 27.8 kDa cellular receptor from flounder (Paralichthys olivaceus). J. Gen. Virol. 2017, 98, 1477–1488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Y.; Ruan, H.M.; Li, Z.Q.; Yuan, X.P.; Gui, J.F. Infection and propagation of lymphocystis virus isolated from the cultured flounder Paralichthys olivaceus in grass carp cell lines. Dis. Aquat. Organ. 2003, 57, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.B.; Cong, R.S.; Fan, T.J.; Mei, X.G. Induction of apoptosis in a flounder gill cell line by lymphocystis disease virus infection. J. Fish Dis. 2004, 27, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.K.; Hijikata, M.; Iwamoto, A.; Alter, H.J.; Purcell, R.H.; Yoshikura, H. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J. Virol. 1994, 68, 1494–1500. [Google Scholar] [PubMed]
- Li, Y.; O’Dell, S.; Walker, L.M.; Wu, X.; Guenaga, J.; Feng, Y.; Schmidt, S.D.; McKee, K.; Louder, M.K.; Ledgerwood, J.E.; et al. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J. Virol. 2011, 85, 8954–8967. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.F.; Zhan, W.B.; Xing, J.; Sheng, X.Z. Development and characterization of monoclonal antibody to the lymphocystis disease virus of Japanese flounder Paralichthys olivaceus isolated from China. J. Virol. Methods 2006, 135, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Sorimachi, M. Production of monoclonal antibodies against red sea bream iridovirus. Fish Pathol. 1995, 30, 47–52. [Google Scholar] [CrossRef]
- Nakajima, K.; Sorimachi, M. Production of monoclonal antibodies against yellowtail ascites virus. J. Fish Dis. 1996, 19, 161–166. [Google Scholar] [CrossRef]
- Zhan, W.B.; Wang, Y.H.; Fryer, J.L.; Okubo, K.; Fukuda, H.; Yu, K.K.; Meng, Q.X. Production of monoclonal antibodies (MAbs) against white spot syndrome virus (WSSV). J. Aquat. Anim. Health 1999, 11, 17–22. [Google Scholar] [CrossRef]
- Poulos, B.T.; Pantoja, C.R.; Bradley-Dunlop, D.; Aguilar, J.; Lightner, D.V. Development and application of monoclonal antibodies for the detection of white spot syndrome virus of penaeid shrimp. Dis. Aquat. Organ. 2001, 47, 13–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gelanew, T.; Hunsperger, E. Development and characterization of serotype specific monoclonal antibodies against the dengue virus-4 (DENV-4) non-structural protein (NS1). Virol. J. 2018, 15, 30. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.T.; Liao, M.Y.; Chiu, C.Y.; Shen, W.F.; Chiu, C.Y.; Cheng, P.C.; Chang, G.J.J.; Wu, H.C. Generation of monoclonal antibodies against dengue virus type 4 and identification of enhancing epitopes on envelope protein. PLoS ONE 2015, 10, e0136328. [Google Scholar] [CrossRef] [PubMed]
- Lentz, T.L. Binding of viral attachment protein to host-cell receptor: The Achilles heel of infectious viruses. Trends Pharmacol. Sci. 1988, 9, 247–252. [Google Scholar] [CrossRef]
- Burton, D.R.; Williamson, R.A.; Paul, W.H.I. Antibody and virus: Binding and neutralization. J. Virol. 2000, 270, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, G.; Correa, I.; Melgosa, M.P.; Bullido, M.J.; Enjuanes, L. Critical epitopes in transmissible gastroenteritis virus neutralization. J. Virol. 1986, 60, 131–139. [Google Scholar] [PubMed]
- Knossow, M.; Gaudier, M.; Douglas, A.; Barrere, B.; Bizebard, T.; Barbey, C.; Gigant, B.; Skehel, J.J. Mechanism of neutralization of influenza virus infectivity by antibodies. J. Virol. 2002, 302, 294–298. [Google Scholar] [CrossRef]
- Wei, X.; Decker, J.M.; Wang, S.; Hui, H.; Kappes, J.C.; Wu, X.Y.; Jesus, F.S.G.; Maria, G.S.; Kilby, J.M.; Michael, S.S.; et al. Antibody neutralization and escape by HIV-1. Nature 2003, 422, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.P.; Dimmock, N.J. Mechanism of neutralization of influenza virus by secretory IgA is different from that of monomeric IgA or IgG. J. Exp. Med. 1985, 161, 198–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiner, H.L.; Fields, B.N. Neutralization of reovirus: The gene responsible for the neutralization antigen. J. Exp. Med. 1977, 146, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.J.; Gong, J.; Huang, X.H.; Huang, Y.H.; Zhou, S.; Ou-Yang, Z.L.; Cao, J.H.; Ye, L.L.; Qin, Q.W. Identification and characterization of a novel capsid protein encoded by Singapore grouper iridovirus ORF038L. Arch. Virol. 2010, 155, 351–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.L.; Zhou, S.; Xia, L.Q.; Huang, X.H.; Huang, Y.H.; Cao, J.H.; Qin, Q.W. Characterization of the VP39 envelope protein from Singapore grouper iridovirus. Can. J. Microbiol. 2015, 61, 924–937. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.B.; Kim, Y.R.; Cha, I.S.; Noh, S.W.; Park, S.B.; Ohtani, M.; Hikima, J.; Aoki, T.; Jung, T.S. Detection of antigenic proteins expressed by lymphocystis virus as vaccine candidates in olive flounder, Paralichthys olivaceus (Temminck & Schlegel). J. Fish Dis. 2011, 34, 555–562. [Google Scholar] [PubMed]
- Keck, Z.Y.; Sung, V.M.; Perkins, S.; Rowe, J.; Paul, S.; Liang, T.J.; Lai, M.C.; Foung, S.K. Human monoclonal antibody to hepatitis C virus E1 glycoprotein that blocks virus attachment and viral infectivity. J. Virol. 2004, 78, 7257–7263. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Wang, L.; Xu, H.; Wang, Z.; Zhang, T.; Wang, M.; Ning, Y.; Deng, F.; Hu, Z.; Wang, H.; et al. A novel glycoprotein D-specific monoclonal antibody neutralizes herpes simplex virus. Antivir. Res. 2017, 147, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.H.; Gong, J.; Huang, Y.H.; Ouyang, Z.L.; Wang, S.W.; Chen, X.L.; Qin, Q.W. Characterization of an envelope gene VP19 from Singapore grouper iridovirus. Virol. J. 2013, 10, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, L.B.; Ke, F.; Wang, J.; Gao, X.C.; Zhang, Q.Y. Rana grylio virus (RGV) envelope protein 2L: Subcellular localization and essential roles in virus infectivity revealed by conditional lethal mutant. J. Gen. Virol. 2014, 95, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Tang, X.Q.; Sheng, X.Z.; Xing, J.; Zhan, W.B. Development and application of monoclonal. antibodies against IgM of black rockfish Sebastes schlegeli. J. Fish Biol. 2017, 90, 1668–1675. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Tang, X.Q.; Sheng, X.Z.; Xing, J.; Zhan, W.B. Development of monoclonal antibodies against IgM of half-smooth tongue sole (Cynoglossus semilaevis) and analysis of phagocytosis of fluorescence microspheres by mIgM+ lymphocytes. Fish Shellfish Immunol. 2017, 66, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Groth, S.F.S. The evaluation of limiting dilution assays. J. Immunol. Methods 1982, 49, R11–R23. [Google Scholar] [CrossRef]
- Wu, R.H.; Tang, X.Q.; Sheng, X.Z.; Xing, J.; Zhan, W.B. Relationship between expression of cellular receptor-27.8 kDa and lymphocystis disease virus (LCDV) infection. PLoS ONE 2015, 10, e0127940. [Google Scholar]
- Perosa, F.; Carbone, R.; Ferrone, S.; Dammacco, F. Purification of human immunoglobulins by sequential precipitation with caprylic acid and ammonium sulphate. J. Immunol. Methods 1990, 128, 9–16. [Google Scholar] [CrossRef]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, Y.; Tang, X.; Sheng, X.; Xing, J.; Zhan, W. Development and Characterization of Monoclonal Antibodies to the 32 kDa Viral Attachment Protein of Lymphocystis Disease Virus and Their Neutralizing Ability in Vitro. Int. J. Mol. Sci. 2018, 19, 2536. https://doi.org/10.3390/ijms19092536
Zhong Y, Tang X, Sheng X, Xing J, Zhan W. Development and Characterization of Monoclonal Antibodies to the 32 kDa Viral Attachment Protein of Lymphocystis Disease Virus and Their Neutralizing Ability in Vitro. International Journal of Molecular Sciences. 2018; 19(9):2536. https://doi.org/10.3390/ijms19092536
Chicago/Turabian StyleZhong, Ying, Xiaoqian Tang, Xiuzhen Sheng, Jing Xing, and Wenbin Zhan. 2018. "Development and Characterization of Monoclonal Antibodies to the 32 kDa Viral Attachment Protein of Lymphocystis Disease Virus and Their Neutralizing Ability in Vitro" International Journal of Molecular Sciences 19, no. 9: 2536. https://doi.org/10.3390/ijms19092536
APA StyleZhong, Y., Tang, X., Sheng, X., Xing, J., & Zhan, W. (2018). Development and Characterization of Monoclonal Antibodies to the 32 kDa Viral Attachment Protein of Lymphocystis Disease Virus and Their Neutralizing Ability in Vitro. International Journal of Molecular Sciences, 19(9), 2536. https://doi.org/10.3390/ijms19092536