Leukocyte Immunoglobulin-Like Receptors A2 and A6 are Expressed in Avian Macrophages and Modulate Cytokine Production by Activating Multiple Signaling Pathways
Abstract
:1. Introduction
2. Results
2.1. Cloning and Identification of Chicken LILRA2 and LILRA6
2.2. Distribution of LILRA2 and LILRA6 Transcripts in Normal Tissues and Cells
2.3. Chicken LILRA2 and LILRA6 Binding with MHC Class I
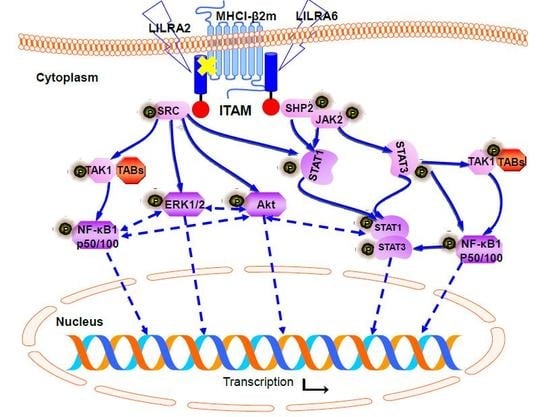
2.4. Chicken LILRA2 and LILRA6 are Associated with Phosphorylation of Src and SHP2
2.5. Chicken LILRA2 and LILRA6 Activate the JAK-STAT Signaling Pathway
2.6. Chicken LILRA2 and LILRA6 Activate NF-κB and MAPK Signaling Pathways
2.7. Chicken LILRA2 and LILRA6 Induce Cytokine Production
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Cloning and Sequencing Analysis of Chicken LILRA2 and LILRA6
4.3. Chicken Tissues and Cell Culture
4.4. Vector Construction and Cell Transfection
4.5. Cytotoxicity Test
4.6. Flow Cytometry
4.7. Immunoprecipitation and Western Blotting
4.8. Enzyme-Linked Immunosorbent Assay (ELISA)
4.9. RNA Extraction and cDNA Synthesis
4.10. Quantitative Reverse Transcription PCR (qRT-PCR)
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NKs | natural killer cells |
| JAK | Janus kinase |
| STAT | signal transducer and activator of transcription |
| IL | interleukin |
| TNF | tumor necrosis factor |
| RT | reverse transcription |
| TYK2 | tyrosine kinase 2 |
| TGFβ | transforming growth factor beta |
| SH2 | Src homology 2-containing tyrosine phosphatase 2 |
| NF-κB | nuclear factor kappa B |
| TAK1 | mitogen-activated protein kinase kinase kinase 7 |
| MAPK | mitogen-activated protein kinases |
| Erk | Extracellular Signal-Regulated Kinase |
| PI3K | Phosphoinositide-3-Kinase |
| JNK | C-Jun N-Terminal Kinase |
| LILR | leukocyte immunoglobulin-like receptor |
| ITAMs | immunoreceptor tyrosine-based activation motifs |
| FcRγ | Fc receptor γ |
References
- Hirayasu, K.; Saito, F.; Suenaga, T.; Shida, K.; Arase, N.; Oikawa, K.; Yamaoka, T.; Murota, H.; Chibana, H.; Nakagawa, I.; et al. Microbially cleaved immunoglobulins are sensed by the innate immune receptor LILRA2. Nat. Microbiol. 2016, 1, 16054. [Google Scholar] [CrossRef] [PubMed]
- Bego, M.G.; Cote, E.; Aschman, N.; Mercier, J.; Weissenhorn, W.; Cohen, E.A. Vpu Exploits the Cross-Talk between BST2 and the ILT7 Receptor to Suppress Anti-HIV-1 Responses by Plasmacytoid Dendritic Cells. PLoS Pathog. 2015, 11, e1005024. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.K.; Mitchell, A.; Endoh, Y.; Hampartzoumian, T.; Huynh, O.; Borges, L.; Geczy, C.; Bryant, K.; Tedla, N. LILRA2 selectively modulates LPS-mediated cytokine production and inhibits phagocytosis by monocytes. PLoS ONE 2012, 7, e33478. [Google Scholar] [CrossRef] [PubMed]
- Jansen, C.A.; van Haarlem, D.A.; Sperling, B.; van Kooten, P.J.; de Vries, E.; Viertlboeck, B.C.; Vervelde, L.; Gobel, T.W. Identification of an Activating Chicken Ig-like Receptor Recognizing Avian Influenza Viruses. J. Immunol. 2016, 197, 4696–4703. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.C.; Hewitt, C.R.; Lopez-Alvarez, M.R.; Jahnke, M.; Russell, A.I.; Radjabova, V.; Trowsdale, A.R.; Trowsdale, J. Allele-specific recognition by LILRB3 and LILRA6 of a cytokeratin 8-associated ligand on necrotic glandular epithelial cells. Oncotarget 2016, 7, 15618–15631. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.J.; Sieling, P.A.; Ochoa, M.T.; Krutzik, S.R.; Guo, B.; Hernandez, M.; Rea, T.H.; Cheng, G.; Colonna, M.; Modlin, R.L. LILRA2 activation inhibits dendritic cell differentiation and antigen presentation to T cells. J. Immunol. 2007, 179, 8128–8136. [Google Scholar] [CrossRef] [PubMed]
- Hirayasu, K.; Arase, H. Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations. Int. J. Hum. Genet. 2015, 60, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Sloane, D.E.; Tedla, N.; Awoniyi, M.; Macglashan, D.W., Jr.; Borges, L.; Austen, K.F.; Arm, J.P. Leukocyte immunoglobulin-like receptors: Novel innate receptors for human basophil activation and inhibition. Blood 2004, 104, 2832–2839. [Google Scholar] [CrossRef] [PubMed]
- Borges, L.; Kubin, M.; Kuhlman, T. LIR9, an immunoglobulin-superfamily-activating receptor, is expressed as a transmembrane and as a secreted molecule. Blood 2003, 101, 1484–1486. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gao, F.; Chu, F.; Peng, H.; Zong, L.; Liu, Y.; Tien, P.; Gao, G.F. Crystal structure of myeloid cell activating receptor leukocyte Ig-like receptor A2 (LILRA2/ILT1/LIR-7) domain swapped dimer: Molecular basis for its non-binding to MHC complexes. J. Mol. Biol. 2009, 386, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Low, H.Z.; Ahrenstorf, G.; Pommerenke, C.; Habermann, N.; Schughart, K.; Ordonez, D.; Stripecke, R.; Wilk, E.; Witte, T. TLR8 regulation of LILRA3 in monocytes is abrogated in human immunodeficiency virus infection and correlates to CD4 counts and virus loads. Retrovirology 2016, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Lim, C.; Guillemin, G.J.; Vollmer-Conna, U.; Rawlinson, W.; Bryant, K.; Tedla, N. Serum Leukocyte Immunoglobulin-Like Receptor A3 (LILRA3) Is Increased in Patients with Multiple Sclerosis and Is a Strong Independent Indicator of Disease Severity; 6.7kbp LILRA3 Gene Deletion Is Not Associated with Diseases Susceptibility. PLoS ONE 2016, 11, e0149200. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Alvarez, M.R.; Jiang, W.; Jones, D.C.; Jayaraman, J.; Johnson, C.; Cookson, W.O.; Moffatt, M.F.; Trowsdale, J.; Traherne, J.A. LILRA6 copy number variation correlates with susceptibility to atopic dermatitis. Immunogenetics 2016, 68, 743–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, A.; Rentero, C.; Endoh, Y.; Hsu, K.; Gaus, K.; Geczy, C.; McNeil, H.P.; Borges, L.; Tedla, N. LILRA5 is expressed by synovial tissue macrophages in rheumatoid arthritis, selectively induces pro-inflammatory cytokines and IL-10 and is regulated by TNF-alpha, IL-10 and IFN-gamma. Eur. J. Immunol. 2008, 38, 3459–3473. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Brettle, M.; Lee, T.; Heng, B.; Lim, C.K.; Guillemin, G.J.; Lord, M.S.; Klotzsch, E.; Geczy, C.L.; Bryant, K.; et al. Soluble LILRA3 promotes neurite outgrowth and synapses formation through a high-affinity interaction with Nogo 66. J. Cell Sci. 2016, 129, 1198–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fei, C.; Pemberton, J.G.; Lillico, D.M.; Zwozdesky, M.A.; Stafford, J.L. Biochemical and Functional Insights into the Integrated Regulation of Innate Immune Cell Responses by Teleost Leukocyte Immune-Type Receptors. Biology 2016, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Montero-Melendez, T.; Llor, X.; Garcia-Planella, E.; Perretti, M.; Suarez, A. Identification of novel predictor classifiers for inflammatory bowel disease by gene expression profiling. PLoS ONE 2013, 8, e76235. [Google Scholar] [CrossRef] [PubMed]
- Tedla, N.; Gibson, K.; McNeil, H.P.; Cosman, D.; Borges, L.; Arm, J.P. The co-expression of activating and inhibitory leukocyte immunoglobulin-like receptors in rheumatoid synovium. Am. J. Pathol. 2002, 160, 425–431. [Google Scholar] [CrossRef]
- Dennis, G., Jr.; Kubagawa, H.; Cooper, M.D. Paired Ig-like receptor homologs in birds and mammals share a common ancestor with mammalian Fc receptors. Proc. Natl. Acad. Sci. USA 2000, 97, 13245–13250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolaidis, N.; Klein, J.; Nei, M. Origin and evolution of the Ig-like domains present in mammalian leukocyte receptors: Insights from chicken, frog and fish homologues. Immunogenetics 2005, 57, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Laun, K.; Coggill, P.; Palmer, S.; Sims, S.; Ning, Z.; Ragoussis, J.; Volpi, E.; Wilson, N.; Beck, S.; Ziegler, A.; et al. The leukocyte receptor complex in chicken is characterized by massive expansion and diversification of immunoglobulin-like Loci. PLoS Genet. 2006, 2, e73. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chu, F.; Gao, F.; Zhou, B.; Gao, G.F. Stability engineering, biophysical and biological characterization of the myeloid activating receptor immunoglobulin-like transcript 1 (ILT1/LIR-7/LILRA2). Protein Expr. Purif. 2007, 56, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Ryu, M.; Chen, Y.; Qi, J.; Liu, J.; Fan, Z.; Nam, G.; Shi, Y.; Cheng, H.; Gao, G.F. LILRA3 binds both classical and non-classical HLA class I molecules but with reduced affinities compared to LILRB1/LILRB2: Structural evidence. PLoS ONE 2011, 6, e19245. [Google Scholar] [CrossRef] [PubMed]
- Shiroishi, M.; Kajikawa, M.; Kuroki, K.; Ose, T.; Kohda, D.; Maenaka, K. Crystal structure of the human monocyte-activating receptor, “Group 2” leukocyte Ig-like receptor A5 (LILRA5/LIR9/ILT11). J. Biol. Chem. 2006, 281, 19536–19544. [Google Scholar] [CrossRef] [PubMed]
- Takai, T. Paired immunoglobulin-like receptors and their MHC class I recognition. Immunology 2005, 115, 433–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeMaoult, J.; Zafaranloo, K.; Le Danff, C.; Carosella, E.D. HLA-G up-regulates ILT2, ILT3, ILT4 and KIR2DL4 in antigen presenting cells, NK cells and T cells. FASEB J. 2005, 19, 662–664. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Nakamura, A. Regulation of immune and neural function via leukocyte Ig-like receptors. J Biochem. 2017, 162, 73–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubagawa, H.; Cooper, M.D.; Chen, C.C.; Ho, L.H.; Alley, T.L.; Hurez, V.; Tun, T.; Uehara, T.; Shimada, T.; Burrows, P.D. Paired immunoglobulin-like receptors of activating and inhibitory types. Curr. Top. Microbiol. Immunol. 1999, 244, 137–149. [Google Scholar] [PubMed]
- Takai, T. Roles of Fc receptors in autoimmunity. Nat. Rev. Immunol. 2002, 2, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Chappell, P.; Meziane el, K.; Harrison, M.; Magiera, L.; Hermann, C.; Mears, L.; Wrobel, A.G.; Durant, C.; Nielsen, L.L.; Buus, S.; et al. Expression levels of MHC class I molecules are inversely correlated with promiscuity of peptide binding. Elife 2015, 4, e05345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Y.; Nathan, D.M.; Li, F.; Li, X.; Faustman, D.L. Defective major histocompatibility complex class I expression on lymphoid cells in autoimmunity. J. Clin. Investig. 1993, 91, 2301–2307. [Google Scholar] [CrossRef] [PubMed]
- Underhill, D.M.; Goodridge, H.S. The many faces of ITAMs. Trends Immunol. 2007, 28, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Mocsai, A.; Ruland, J.; Tybulewicz, V.L. The SYK tyrosine kinase: A crucial player in diverse biological functions. Nat. Rev. Immunol. 2010, 10, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Q.; Yang, W.; Kontaridis, M.I.; Bivona, T.G.; Wen, G.; Araki, T.; Luo, J.; Thompson, J.A.; Schraven, B.L.; Philips, M.R.; et al. Shp2 regulates SRC family kinase activity and Ras/Erk activation by controlling Csk recruitment. Mol. Cell 2004, 13, 341–355. [Google Scholar] [CrossRef]
- Fodor, S.; Jakus, Z.; Mocsai, A. ITAM-based signaling beyond the adaptive immune response. Immunol. Lett. 2006, 104, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Abram, C.L.; Lowell, C.A. The diverse functions of Src family kinases in macrophages. Front Biosci. 2008, 13, 4426–4450. [Google Scholar] [CrossRef] [PubMed]
- Nelin, L.D.; White, H.A.; Jin, Y.; Trittmann, J.K.; Chen, B.; Liu, Y. The Src family tyrosine kinases src and yes have differential effects on inflammation-induced apoptosis in human pulmonary microvascular endothelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2016, 310, L880–L888. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dillmann, C.; Ringel, C.; Ringleb, J.; Mora, J.; Olesch, C.; Fink, A.F.; Roberts, E.; Brune, B.; Weigert, A. S1PR4 Signaling Attenuates ILT 7 Internalization To Limit IFN-alpha Production by Human Plasmacytoid Dendritic Cells. J. Immunol. 2016, 196, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Steelman, L.S.; Pohnert, S.C.; Shelton, J.G.; Franklin, R.A.; Bertrand, F.E.; McCubrey, J.A. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia 2004, 18, 189–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vakkila, J.; Demarco, R.A.; Lotze, M.T. Coordinate NF-kappaB and STAT1 activation promotes development of myeloid type 1 dendritic cells. Scand. J. Immunol. 2008, 67, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Burke, S.J.; Lu, D.; Sparer, T.E.; Masi, T.; Goff, M.R.; Karlstad, M.D.; Collier, J.J. NF-kappaB and STAT1 control CXCL1 and CXCL2 gene transcription. Am. J. Physiol. Endocrinol. Metabol. 2014, 306, E131–E149. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Mao, R.; Yang, J. NF-kappaB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013, 4, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Kojima, H.; Sasaki, T.; Ishitani, T.; Iemura, S.; Zhao, H.; Kaneko, S.; Kunimoto, H.; Natsume, T.; Matsumoto, K.; Nakajima, K. STAT3 regulates Nemo-like kinase by mediating its interaction with IL-6-stimulated TGFbeta-activated kinase 1 for STAT3 Ser-727 phosphorylation. Proc. Natl. Acad. Sci. USA 2005, 102, 4524–4529. [Google Scholar] [CrossRef] [PubMed]
- Ohkawara, B.; Shirakabe, K.; Hyodo-Miura, J.; Matsuo, R.; Ueno, N.; Matsumoto, K.; Shibuya, H. Role of the TAK1-NLK-STAT3 pathway in TGF-beta-mediated mesoderm induction. Genes Dev. 2004, 18, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Tengvall, S.; Che, K.F.; Linden, A. Interleukin-26: An Emerging Player in Host Defense and Inflammation. J. Innate Immun. 2016, 8, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Truong, A.D.; Park, B.; Ban, J.; Hong, Y.H. The novel chicken interleukin 26 protein is overexpressed in T cells and induces proinflammatory cytokines. BMC Vet. Res. 2016, 47, 65. [Google Scholar] [CrossRef] [PubMed]
- Truong, A.D.; Ban, J.; Park, B.; Hong, Y.H.; Lillehoj, H.S. Characterization and functional analyses of a novel chicken CD8alpha variant X1 (CD8alpha1). J. Anim. Sci. 2016, 94, 2737–2751. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, A.; Chang, H.Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic. Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef] [PubMed]
- Schat, K.A.; Pratt, W.D.; Morgan, R.; Weinstock, D.; Calnek, B.W. Stable transfection of reticuloendotheliosis virus-transformed lymphoblastoid cell lines. Avian Dis. 1992, 36, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Klasing, K.C.; Peng, R.K. Influence of cell sources, stimulating agents and incubation conditions on release of interleukin-1 from chicken macrophages. Dev. Comp. Immunol. 1987, 11, 385–394. [Google Scholar] [CrossRef]
- Winding, P.; Berchtold, M.W. The chicken B cell line DT40: A novel tool for gene disruption experiments. J. Immunol. Methods 2001, 249, 1–16. [Google Scholar] [CrossRef]
- Rocha, E.M.; Hollingdale, M.R.; Gwadz, R.; Krettli, A.U. Exoerythrocytic development of Plasmodium gallinaceum sporozoites in a chicken fibroblast cell line and inhibition of the cell invasion by specific anti-sporozoite monoclonal antibodies. J. Eukaryot. Microbiol. 1993, 40, 64–66. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, W.; Witter, R.L.; Romero, C.; Nazerian, K.; Sharma, J.M.; Fadly, A.; Ewert, D. Induction of lymphoid leukosis transplant able tumours and the establishment of lymphoblastoid cell lines. Avian Pathol. 1980, 9, 311–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truong, A.D.; Hong, Y.; Hoang, C.T.; Lee, J.; Hong, Y.H. Chicken IL-26 regulates immune responses through the JAK/STAT and NF-kappaB signaling pathways. Dev. Comp. Immunol. 2017, 73, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]









| Genes | Species | LILRA2 | LILRA6 | LILRA2 | LILRA6 | LILRA2 | LILRA6 | LILRA2 | LILRA6 | LILRA2 | LILRA6 | GenBank Acc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chicken | Human | Monkey | Chimpanzee | Pig | ||||||||
| LILRA2 | Chicken | 50.08 | 38.97 | 39.50 | 27.86 | 39.15 | 38.97 | 38.97 | 55.37 | 39.15 | XP_004949869 | |
| LILRA6 | 66.13 | 47.08 | 48.32 | 36.68 | 46.20 | 46.56 | 47.44 | 59.78 | 46.91 | XP_003643922 | ||
| LILRA2 | Human | 20.81 | 28.57 | 83.77 | 78.83 | 78.13 | 98.58 | 83.42 | 59.25 | 78.48 | NP_001124389 | |
| LILRA6 | 20.45 | 28.57 | 72.83 | 71.25 | 83.77 | 83.24 | 94.53 | 60.14 | 82.36 | NP_077294 | ||
| LILRA2 | Monkey | 19.70 | 17.81 | 70.37 | 58.02 | 68.95 | 78.13 | 70.54 | 46.03 | 65.25 | NP_001035761 | |
| LILRA6 | 20.98 | 27.33 | 67.54 | 75.66 | 55.55 | 78.13 | 83.95 | 56.26 | 76.54 | NP_001035764 | ||
| LILRA2 | Chimpanzee | 20.45 | 28.74 | 96.64 | 73.01 | 70.37 | 67.54 | 83.24 | 59.08 | 77.95 | XP_009434592 | |
| LILRA6 | 20.81 | 28.39 | 73.19 | 90.82 | 56.96 | 76.89 | 73.36 | 61.19 | 81.83 | NP_001009056 | ||
| LILRA2 | Pig | 37.56 | 44.26 | 45.50 | 46.73 | 31.04 | 43.03 | 45.14 | 47.26 | 64.02 | XP_013846133 | |
| LILRA6 | 20.63 | 26.63 | 63.49 | 69.31 | 50.08 | 63.49 | 64.02 | 69.48 | 49.91 | XP_003134221 | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Truong, A.D.; Rengaraj, D.; Hong, Y.; Tran, H.T.T.; Dang, H.V.; Nguyen, V.K.; Lillehoj, H.S.; Hong, Y.H. Leukocyte Immunoglobulin-Like Receptors A2 and A6 are Expressed in Avian Macrophages and Modulate Cytokine Production by Activating Multiple Signaling Pathways. Int. J. Mol. Sci. 2018, 19, 2710. https://doi.org/10.3390/ijms19092710
Truong AD, Rengaraj D, Hong Y, Tran HTT, Dang HV, Nguyen VK, Lillehoj HS, Hong YH. Leukocyte Immunoglobulin-Like Receptors A2 and A6 are Expressed in Avian Macrophages and Modulate Cytokine Production by Activating Multiple Signaling Pathways. International Journal of Molecular Sciences. 2018; 19(9):2710. https://doi.org/10.3390/ijms19092710
Chicago/Turabian StyleTruong, Anh Duc, Deivendran Rengaraj, Yeojin Hong, Ha Thi Thanh Tran, Hoang Vu Dang, Viet Khong Nguyen, Hyun S. Lillehoj, and Yeong Ho Hong. 2018. "Leukocyte Immunoglobulin-Like Receptors A2 and A6 are Expressed in Avian Macrophages and Modulate Cytokine Production by Activating Multiple Signaling Pathways" International Journal of Molecular Sciences 19, no. 9: 2710. https://doi.org/10.3390/ijms19092710
APA StyleTruong, A. D., Rengaraj, D., Hong, Y., Tran, H. T. T., Dang, H. V., Nguyen, V. K., Lillehoj, H. S., & Hong, Y. H. (2018). Leukocyte Immunoglobulin-Like Receptors A2 and A6 are Expressed in Avian Macrophages and Modulate Cytokine Production by Activating Multiple Signaling Pathways. International Journal of Molecular Sciences, 19(9), 2710. https://doi.org/10.3390/ijms19092710