Polybenzyl Glutamate Biocompatible Scaffold Promotes the Efficiency of Retinal Differentiation toward Retinal Ganglion Cell Lineage from Human-Induced Pluripotent Stem Cells
Abstract
:1. Introduction
2. Results
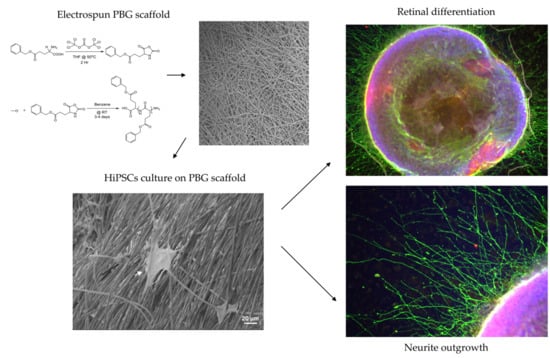
2.1. Induction of hiPSC Differentiation to RGC-Like Cells on PBG Scaffold
2.2. Determination of RGC Lineages and Axon Growth of hiPSC-Derived RGC-Like Cells
2.3. RNA-seq Analysis of iPSC-Derived RGC-Like Cells Cultured on PBG Scaffold
3. Discussion
4. Materials and Methods
4.1. Human iPSC (hiPSC) Culture
4.2. Induction of hiPSC Differentiation to RGCs
4.3. Electrospun PBG Scaffolds
4.4. Real-Time PCR
4.5. Immunofluorescence Staining
4.6. Measurement of Neurites
4.7. SEM Observation
4.8. Whole Transcriptome Analysis by RNA-Seq
4.9. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Availability of Data and Materials
Conflicts of Interest
References
- Quigley, H.A.; Broman, A.T. The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 2006, 90, 262–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansour-Robaey, S.; Clarke, D.B.; Wang, Y.C.; Bray, G.M.; Aguayo, A.J. Effects of ocular injury and administration of brain-derived neurotrophic factor on survival and regrowth of axotomized retinal ganglion cells. Proc. Natl. Acad. Sci. USA 1994, 91, 1632–1636. [Google Scholar] [CrossRef] [PubMed]
- Park, K.K.; Liu, K.; Hu, Y.; Smith, P.D.; Wang, C.; Cai, B.; Xu, B.; Connolly, L.; Kramvis, I.; Sahin, M.; et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 2008, 322, 963–966. [Google Scholar] [CrossRef] [PubMed]
- Baltmr, A.; Duggan, J.; Nizari, S.; Salt, T.E.; Cordeiro, M.F. Neuroprotection in glaucoma—Is there a future role? Exp. Eye Res. 2010, 91, 554–566. [Google Scholar] [CrossRef] [PubMed]
- Danesh-Meyer, H.V.; Levin, L.A. Neuroprotection: Extrapolating from neurologic diseases to the eye. Am. J. Ophthalmol. 2009, 148, 186–191.e182. [Google Scholar] [CrossRef] [PubMed]
- Dahlmann-Noor, A.H.; Vijay, S.; Limb, G.A.; Khaw, P.T. Strategies for optic nerve rescue and regeneration in glaucoma and other optic neuropathies. Drug Discov. Today 2010, 15, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Osakada, F.; Ikeda, H.; Mandai, M.; Wataya, T.; Watanabe, K.; Yoshimura, N.; Akaike, A.; Sasai, Y.; Takahashi, M. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat. Biotechnol. 2008, 26, 215–224. [Google Scholar] [CrossRef]
- Buchholz, D.E.; Hikita, S.T.; Rowland, T.J.; Friedrich, A.M.; Hinman, C.R.; Johnson, L.V.; Clegg, D.O. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells 2009, 27, 2427–2434. [Google Scholar] [CrossRef]
- Geng, Z.; Walsh, P.J.; Truong, V.; Hill, C.; Ebeling, M.; Kapphahn, R.J.; Montezuma, S.R.; Yuan, C.; Roehrich, H.; Ferrington, D.A.; et al. Generation of retinal pigmented epithelium from iPSCs derived from the conjunctiva of donors with and without age related macular degeneration. PLoS ONE 2017, 12, e0173575. [Google Scholar] [CrossRef]
- Ohlemacher, S.K.; Sridhar, A.; Xiao, Y.; Hochstetler, A.E.; Sarfarazi, M.; Cummins, T.R.; Meyer, J.S. Stepwise Differentiation of Retinal Ganglion Cells from Human Pluripotent Stem Cells Enables Analysis of Glaucomatous Neurodegeneration. Stem Cells 2016, 34, 1553–1562. [Google Scholar] [CrossRef] [Green Version]
- Osakada, F.; Ikeda, H.; Sasai, Y.; Takahashi, M. Stepwise differentiation of pluripotent stem cells into retinal cells. Nat. Protoc. 2009, 4, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Mesentier-Louro, L.A.; Zaverucha-do-Valle, C.; da Silva-Junior, A.J.; Nascimento-Dos-Santos, G.; Gubert, F.; de Figueirêdo, A.B.; Torres, A.L.; Paredes, B.D.; Teixeira, C.; Tovar-Moll, F.; et al. Distribution of mesenchymal stem cells and effects on neuronal survival and axon regeneration after optic nerve crush and cell therapy. PLoS ONE 2014, 9, e110722. [Google Scholar] [CrossRef]
- Cho, J.H.; Mao, C.A.; Klein, W.H. Adult mice transplanted with embryonic retinal progenitor cells: New approach for repairing damaged optic nerves. Mol. Vis. 2012, 18, 2658–2672. [Google Scholar] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Okita, K.; Ichisaka, T.; Yamanaka, S. Generation of germline-competent induced pluripotent stem cells. Nature 2007, 448, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Cuzzani, O.; Binette, F.; Sternberg, H.; West, M.D.; Nasonkin, I.O. Pluripotent Stem Cells for Retinal Tissue Engineering: Current Status and Future Prospects. Stem Cell Rev. 2018, 14, 463–483. [Google Scholar] [CrossRef]
- Tanaka, T.; Yokoi, T.; Tamalu, F.; Watanabe, S.; Nishina, S.; Azuma, N. Generation of retinal ganglion cells with functional axons from human induced pluripotent stem cells. Sci. Rep. 2015, 5, 8344. [Google Scholar] [CrossRef] [Green Version]
- Álvarez, Z.; Castaño, O.; Castells, A.A.; Mateos-Timoneda, M.A.; Planell, J.A.; Engel, E.; Alcántara, S. Neurogenesis and vascularization of the damaged brain using a lactate-releasing biomimetic scaffold. Biomaterials 2014, 35, 4769–4781. [Google Scholar] [CrossRef]
- Orive, G.; Anitua, E.; Pedraz, J.L.; Emerich, D.F. Biomaterials for promoting brain protection, repair and regeneration. Nat. Rev. Neurosci. 2009, 10, 682–692. [Google Scholar] [CrossRef]
- Hench, L.L.; Polak, J.M. Third-generation biomedical materials. Science 2002, 295, 1014–1017. [Google Scholar] [CrossRef] [PubMed]
- Simitzi, C.; Ranella, A.; Stratakis, E. Controlling the morphology and outgrowth of nerve and neuroglial cells: The effect of surface topography. Acta Biomater. 2017, 51, 21–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.H.; Chang, Y.Y.; Wu, J.G.; Lin, C.Y.; An, H.L.; Luo, S.C.; Tang, T.K.; Su, W.F. Novel 3D Neuron Regeneration Scaffolds Based on Synthetic Polypeptide Containing Neuron Cue. Macromol. Biosci. 2018, 18, 1700251–1700263. [Google Scholar] [CrossRef] [PubMed]
- Kador, K.E.; Alsehli, H.S.; Zindell, A.N.; Lau, L.W.; Andreopoulos, F.M.; Watson, B.D.; Goldberg, J.L. Retinal ganglion cell polarization using immobilized guidance cues on a tissue-engineered scaffold. Acta Biomater. 2014, 10, 4939–4946. [Google Scholar] [CrossRef] [Green Version]
- Kador, K.E.; Montero, R.B.; Venugopalan, P.; Hertz, J.; Zindell, A.N.; Valenzuela, D.A.; Uddin, M.S.; Lavik, E.B.; Muller, K.J.; Andreopoulos, F.M.; et al. Tissue engineering the retinal ganglion cell nerve fiber layer. Biomaterials 2013, 34, 4242–4250. [Google Scholar] [CrossRef] [Green Version]
- Aizawa, Y.; Shoichet, M.S. The role of endothelial cells in the retinal stem and progenitor cell niche within a 3D engineered hydrogel matrix. Biomaterials 2012, 33, 5198–5205. [Google Scholar] [CrossRef]
- Kuo, Y.C.; Huang, M.J. Material-driven differentiation of induced pluripotent stem cells in neuron growth factor-grafted poly(ε-caprolactone)-poly(β-hydroxybutyrate) scaffolds. Biomaterials 2012, 33, 5672–5682. [Google Scholar] [CrossRef]
- Iwasaki, M.; Wilcox, J.T.; Nishimura, Y.; Zweckberger, K.; Suzuki, H.; Wang, J.; Liu, Y.; Karadimas, S.K.; Fehlings, M.G. Synergistic effects of self-assembling peptide and neural stem/progenitor cells to promote tissue repair and forelimb functional recovery in cervical spinal cord injury. Biomaterials 2014, 35, 2617–2629. [Google Scholar] [CrossRef]
- Prabhakaran, M.P.; Venugopal, J.R.; Ramakrishna, S. Mesenchymal stem cell differentiation to neuronal cells on electrospun nanofibrous substrates for nerve tissue engineering. Biomaterials 2009, 30, 4996–5003. [Google Scholar] [CrossRef]
- Sun, Y.; Li, W.; Wu, X.; Zhang, N.; Zhang, Y.; Ouyang, S.; Song, X.; Fang, X.; Seeram, R.; Xue, W.; et al. Functional Self-Assembling Peptide Nanofiber Hydrogels Designed for Nerve Degeneration. ACS Appl. Mater. Interfaces 2016, 8, 2348–2359. [Google Scholar] [CrossRef]
- Silva, G.A.; Czeisler, C.; Niece, K.L.; Beniash, E.; Harrington, D.A.; Kessler, J.A.; Stupp, S.I. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science 2004, 303, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M.; Takamiya, A.; Jiao, J.W.; Cho, K.S.; Trevino, S.G.; Matsuda, T.; Chen, D.F. alpha-Aminoadipate induces progenitor cell properties of Müller glia in adult mice. Invest. Ophthalmol. Vis. Sci. 2008, 49, 1142–1150. [Google Scholar] [CrossRef] [PubMed]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef]
- Green, E.S.; Stubbs, J.L.; Levine, E.M. Genetic rescue of cell number in a mouse model of microphthalmia: Interactions between Chx10 and G1-phase cell cycle regulators. Development 2003, 130, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Rowan, S.; Cepko, C.L. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev. Biol. 2004, 271, 388–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moshiri, A.; Gonzalez, E.; Tagawa, K.; Maeda, H.; Wang, M.; Frishman, L.J.; Wang, S.W. Near complete loss of retinal ganglion cells in the math5/brn3b double knockout elicits severe reductions of other cell types during retinal development. Dev. Biol. 2008, 316, 214–227. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Morrow, E.M.; Cepko, C.L. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 1997, 91, 531–541. [Google Scholar] [CrossRef]
- Freund, C.L.; Gregory-Evans, C.Y.; Furukawa, T.; Papaioannou, M.; Looser, J.; Ploder, L.; Bellingham, J.; Ng, D.; Herbrick, J.A.; Duncan, A.; et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 1997, 91, 543–553. [Google Scholar] [CrossRef]
- Fuhrmann, S. Eye morphogenesis and patterning of the optic vesicle. Curr. Top. Dev. Biol. 2010, 93, 61–84. [Google Scholar] [CrossRef]
- Sharma, R.K.; Netland, P.A. Early born lineage of retinal neurons express class III beta-tubulin isotype. Brain Res. 2007, 1176, 11–17. [Google Scholar] [CrossRef]
- Choi, J.; Lee, S.; Mallard, W.; Clement, K.; Tagliazucchi, G.M.; Lim, H.; Choi, I.Y.; Ferrari, F.; Tsankov, A.M.; Pop, R.; et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat. Biotechnol. 2015, 33, 1173–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farkas, M.H.; Grant, G.R.; White, J.A.; Sousa, M.E.; Consugar, M.B.; Pierce, E.A. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genom. 2013, 14, 486. [Google Scholar] [CrossRef] [PubMed]
- Kamao, H.; Mandai, M.; Okamoto, S.; Sakai, N.; Suga, A.; Sugita, S.; Kiryu, J.; Takahashi, M. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Rep. 2014, 2, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Mandai, M.; Watanabe, A.; Kurimoto, Y.; Hirami, Y.; Morinaga, C.; Daimon, T.; Fujihara, M.; Akimaru, H.; Sakai, N.; Shibata, Y.; et al. Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration. N. Engl. J. Med. 2017, 376, 1038–1046. [Google Scholar] [CrossRef] [PubMed]
- Erskine, L.; Herrera, E. Connecting the retina to the brain. ASN Neuro 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Chamling, X.; Sluch, V.M.; Zack, D.J. The Potential of Human Stem Cells for the Study and Treatment of Glaucoma. Invest. Ophthalmol. Vis. Sci. 2016, 57, ORSFi1-6. [Google Scholar] [CrossRef]
- Lamba, D.A.; Karl, M.O.; Ware, C.B.; Reh, T.A. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2006, 103, 12769–12774. [Google Scholar] [CrossRef] [Green Version]
- Gill, K.P.; Hung, S.S.; Sharov, A.; Lo, C.Y.; Needham, K.; Lidgerwood, G.E.; Jackson, S.; Crombie, D.E.; Nayagam, B.A.; Cook, A.L.; et al. Enriched retinal ganglion cells derived from human embryonic stem cells. Sci. Rep. 2016, 6, 30552. [Google Scholar] [CrossRef]
- Sluch, V.M.; Davis, C.H.; Ranganathan, V.; Kerr, J.M.; Krick, K.; Martin, R.; Berlinicke, C.A.; Marsh-Armstrong, N.; Diamond, J.S.; Mao, H.Q.; et al. Differentiation of human ESCs to retinal ganglion cells using a CRISPR engineered reporter cell line. Sci. Rep. 2015, 5, 16595. [Google Scholar] [CrossRef] [Green Version]
- Maekawa, Y.; Onishi, A.; Matsushita, K.; Koide, N.; Mandai, M.; Suzuma, K.; Kitaoka, T.; Kuwahara, A.; Ozone, C.; Nakano, T.; et al. Optimized Culture System to Induce Neurite Outgrowth From Retinal Ganglion Cells in Three-Dimensional Retinal Aggregates Differentiated From Mouse and Human Embryonic Stem Cells. Curr. Eye Res. 2016, 41, 558–568. [Google Scholar] [CrossRef]
- Kobayashi, W.; Onishi, A.; Tu, H.Y.; Takihara, Y.; Matsumura, M.; Tsujimoto, K.; Inatani, M.; Nakazawa, T.; Takahashi, M. Culture Systems of Dissociated Mouse and Human Pluripotent Stem Cell-Derived Retinal Ganglion Cells Purified by Two-Step Immunopanning. Investig. Ophthalmol. Vis. Sci. 2018, 59, 776–787. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Ando, S.; Takata, N.; Kawada, M.; Muguruma, K.; Sekiguchi, K.; Saito, K.; Yonemura, S.; Eiraku, M.; Sasai, Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 2012, 10, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Willard, S.S.; Koochekpour, S. Glutamate, glutamate receptors, and downstream signaling pathways. Int. J. Biol. Sci. 2013, 9, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Dhandayuthapani, B.; Yoshida, Y.; Maekawa, T.; Kumar, D.S. Polymeric scaffolds in tissue engineering application: A review. Int. J. Polym. Sci. 2011, 2011, 290602. [Google Scholar] [CrossRef]
- Laughter, M.R.; Ammar, D.A.; Bardill, J.R.; Pena, B.; Kahook, M.Y.; Lee, D.J.; Park, D. A self injectable biomimetic microenvironment encourages retinal ganglion cell axon extension in vitro. ACS Appl. Mater. Interfaces 2016, 8, 20540–20548. [Google Scholar] [CrossRef] [PubMed]
- Gill, K.P.; Hewitt, A.W.; Davidson, K.C.; Pébay, A.; Wong, R.C. Methods of Retinal Ganglion Cell Differentiation From Pluripotent Stem Cells. Transl. Vis. Sci. Technol. 2014, 3, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerber, U. Metabotropic glutamate receptors in vertebrate retina. Doc. Ophthalmol. 2003, 106, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Thoreson, W.B.; Witkovsky, P. Glutamate receptors and circuits in the vertebrate retina. Prog. Retin. Eye Res. 1999, 18, 765–810. [Google Scholar] [CrossRef]
- Connaughton, V. Glutamate and Glutamate Receptors in the Vertebrate Retina. In Webvision: The Organization of the Retina and Visual System; Kolb, H., Fernandez, E., Nelson, R., Eds.; University of Utah Health Sciences Center: Salt Lake City, UT, USA, 1995. [Google Scholar]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Wu, C.E.; Yu, C.W.; Chang, K.W.; Chou, W.H.; Lu, C.Y.; Ghelfi, E.; Wu, F.C.; Jan, P.S.; Huang, M.C.; Allard, P.; et al. Comparative global immune-related gene profiling of somatic cells, human pluripotent stem cells and their derivatives: Implication for human lymphocyte proliferation. Exp. Mol. Med. 2017, 49, e376. [Google Scholar] [CrossRef]
- Chen, P.-H.; Liao, H.-C.; Hsu, S.-H.; Chen, R.-S.; Wu, M.-C.; Yang, Y.-F.; Wu, C.-C.; Chen, M.-H.; Su, W.-F. A novel polyurethane/cellulose fibrous scaffold for cardiac tissue engineering. RSC Adv. 2015, 5, 6932–6939. [Google Scholar] [CrossRef]
- Lan, C.W.; Chen, M.J.; Jan, P.S.; Chen, H.F.; Ho, H.N. Differentiation of human embryonic stem cells into functional ovarian granulosa-like cells. J. Clin. Endocrinol. Metab. 2013, 98, 3713–3723. [Google Scholar] [CrossRef] [PubMed]







| Biological Process (Gene Ontology) | Fold Enrichment | FDR |
|---|---|---|
| Phototransduction (GO:0007602) | 3.45 | 1.35 × 10−4 |
| Photoreceptor cell development (GO:0042462) | 3.16 | 7.53 × 10−3 |
| Retinol metabolic process (GO:0042572) | 3.08 | 1.29 × 10−2 |
| Photoreceptor cell differentiation (GO:0046530) | 2.66 | 4.59 × 10−3 |
| Eye morphogenesis (GO:0048592) | 1.94 | 4.53 × 10−3 |
| Eye development (GO:0001654) | 1.91 | 1.09 × 10−6 |
| Retina development (GO:0060041) | 1.83 | 2.72 × 10−2 |
| Gene | Accession No. | Forward | Reverse |
|---|---|---|---|
| Hprt1 | NM_000194.2 | GGCAGTATAATCCAAAGATGGTCAA | GTCAAGGGCATATCCTACAACAAAC |
| Chx10 | NM_182894.2 | AACCCAATCTGGCTGGTAAATGA | CAGCAGGCCCTTAATGCGTA |
| Brn3b | NM_004575.2 | TGACACATGAGCGCTCTCACTTAC | ACCAAGTGGCAAATGCACCTA |
| Crx | NM_000554.4 | ACCCTGATCTCTAGAGCCCACAA | CTTAATGTCCCAGAACCCAGCA |
| Math5 | NM_145178.3 | CCCTAAATTTGGGCAAGTGAAGA | CAAAGCAACTCACGTGCAATC |
| Tuj1 | NM_006086.3 | GGCCAAGGGTCACTACACG | GCAGTCGCAGTTTTCACACTC |
| Tau | NM_001123066.3 | CCAAGTGTGGCTCATTAGGCA | CCAATCTTCGACTGGACTCTGT |
| NFM | NM_005382.2 | ACAACCACGACCTCAGCAGCTA | ATGACGAGCCATTTCCCACTTT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.-C.; She, P.-Y.; Chen, D.F.; Lu, J.-H.; Yang, C.-H.; Huang, D.-S.; Chen, P.-Y.; Lu, C.-Y.; Cho, K.-S.; Chen, H.-F.; et al. Polybenzyl Glutamate Biocompatible Scaffold Promotes the Efficiency of Retinal Differentiation toward Retinal Ganglion Cell Lineage from Human-Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2019, 20, 178. https://doi.org/10.3390/ijms20010178
Chen T-C, She P-Y, Chen DF, Lu J-H, Yang C-H, Huang D-S, Chen P-Y, Lu C-Y, Cho K-S, Chen H-F, et al. Polybenzyl Glutamate Biocompatible Scaffold Promotes the Efficiency of Retinal Differentiation toward Retinal Ganglion Cell Lineage from Human-Induced Pluripotent Stem Cells. International Journal of Molecular Sciences. 2019; 20(1):178. https://doi.org/10.3390/ijms20010178
Chicago/Turabian StyleChen, Ta-Ching, Pin-Yi She, Dong Feng Chen, Jui-Hsien Lu, Chang-Hao Yang, Ding-Siang Huang, Pao-Yang Chen, Chen-Yu Lu, Kin-Sang Cho, Hsin-Fu Chen, and et al. 2019. "Polybenzyl Glutamate Biocompatible Scaffold Promotes the Efficiency of Retinal Differentiation toward Retinal Ganglion Cell Lineage from Human-Induced Pluripotent Stem Cells" International Journal of Molecular Sciences 20, no. 1: 178. https://doi.org/10.3390/ijms20010178
APA StyleChen, T. -C., She, P. -Y., Chen, D. F., Lu, J. -H., Yang, C. -H., Huang, D. -S., Chen, P. -Y., Lu, C. -Y., Cho, K. -S., Chen, H. -F., & Su, W. -F. (2019). Polybenzyl Glutamate Biocompatible Scaffold Promotes the Efficiency of Retinal Differentiation toward Retinal Ganglion Cell Lineage from Human-Induced Pluripotent Stem Cells. International Journal of Molecular Sciences, 20(1), 178. https://doi.org/10.3390/ijms20010178