Synthetic Tyrosine tRNA Molecules with Noncanonical Secondary Structures
Abstract
:1. Introduction
2. Results
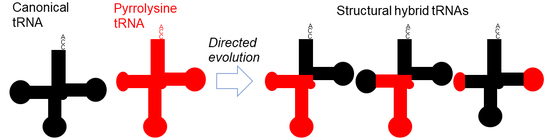
2.1. Hybrid Molecules between tRNAPyl and tRNATyr
2.2. Selections 1 and 2: Sequence Variations in the auD Helix
2.3. Selection 3: Sequence Variation in the D Loop against the T Loop of Mm tRNAPyl
2.4. Selection 4: Sequence Covariation between the D and T Loops in PYLY1 and GGCT1 (ZtRNA)
2.5. Selection 5: Selection of Structural Hybrids having the Canonical auD Helix and Noncanonical Interloop Interactions (YtRNA)
2.6. Selection 6: ZtRNA Scaffold Possibly Supports Glycylation and Glutaminylation in E. coli
3. Discussion
4. Materials and Methods
4.1. Amino Acid and E. coli Strains
4.2. tRNA Genes
4.3. Plasmids
4.4. In Vivo tRNA Activity Assay
4.5. Isolation of tRNA Variants
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| auD | Augmented D |
| aaRS | Aminoacyl-tRNA synthetase |
| TyrRS | Tyrosyl-tRNA synthetase |
| GlyRS | Glycyl-tRNA synthetase |
| GlnRS | Glutaminyl-tRNA synthetase |
References
- Kim, S.H.; Suddath, F.L.; Quigley, G.J.; McPherson, A.; Sussman, J.L.; Wang, A.H.; Seeman, N.C.; Rich, A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 1974, 185, 435–440. [Google Scholar] [CrossRef]
- Ladner, J.E.; Jack, A.; Robertus, J.D.; Brown, R.S.; Rhodes, D.; Clark, B.F.; Klug, A. Structure of yeast phenylalanine transfer RNA at 2.5 Å resolution. Proc. Natl. Acad. Sci. USA 1975, 72, 4414–4418. [Google Scholar] [CrossRef]
- Dirheimer, G.; Keith, G.; Dumas, P.; Westhof, E. Primary, secondary, and tertiary structures of tRNAs. In tRNA: Structure, Biosynthesis, and Function; Söll, D., RajBhandary, U.L., Eds.; AMS Press: Washington, DC, USA, 1995; pp. 93–126. ISBN 155581073X. [Google Scholar]
- Tharp, J.M.; Ehnbom, A.; Liu, W.R. tRNAPyl: Structure, function, and applications. RNA Biol. 2017, 24, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Vargas-Rodriguez, O.; Englert, M.; Tripp, H.J.; Ivanova, N.N.; Rubin, E.M.; Kyrpides, N.C.; Söll, D. Transfer RNAs with novel cloverleaf structures. Nucleic Acids Res. 2017, 45, 2776–2785. [Google Scholar] [CrossRef] [PubMed]
- Normanly, J.; Ogden, R.C.; Horvath, S.J.; Abelson, J. Changing the identity of a transfer RNA. Nature 1986, 321, 213–219. [Google Scholar] [CrossRef]
- Biou, V.; Yaremchuk, A.; Tukalo, M.; Cusack, S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science 1994, 263, 1404–1410. [Google Scholar] [CrossRef]
- Hauenstein, S.; Zhang, C.M.; Hou, Y.M.; Perona, J.J. Shape-selective RNA recognition by cysteinyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2004, 11, 1134–1141. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, R.; Yokoyama, S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol. 2005, 12, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Tukalo, M.; Yaremchuk, A.; Fukunaga, R.; Yokoyama, S.; Cusack, S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat. Struct. Mol. Biol. 2005, 12, 923–930. [Google Scholar] [CrossRef]
- Nozawa, K.; O’Donoghue, P.; Gundllapalli, S.; Araiso, Y.; Ishitani, R.; Umehara, T.; Söll, D.; Nureki, O. Pyrrolysyl-tRNA synthetase–tRNAPyl structure reveals the molecular basis of orthogonality. Nature 2009, 457, 1163–1167. [Google Scholar] [CrossRef]
- Suzuki, T.; Miller, C.; Guo, L.T.; Ho, J.M.L.; Bryson, D.I.; Wang, Y.S.; Liu, D.R.; Söll, D. Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase. Nat. Chem. Biol. 2017, 13, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, K.S.; Crnković, A.; Söll, D. Versatility of synthetic tRNAs in genetic code expansion. Genes 2018, 9, 537. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Murayama, K.; Oki, K.; Iraha, F.; Kato-Murayama, M.; Takahashi, M.; Ohtake, K.; Kobayashi, T.; Kuramitsu, S.; Shirouzu, M.; et al. Genetic encoding of 3-iodo-l-tyrosine in Escherichia coli for single-wavelength anomalous dispersion phasing in protein crystallography. Structure 2009, 17, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Brock, A.; Herberich, B.; Schultz, P.G. Expanding the genetic code of Escherichia coli. Science 2001, 292, 498–500. [Google Scholar] [CrossRef]
- Wang, L.; Magliery, T.J.; Liu, D.R.; Schultz, P.G. A new functional suppressor tRNA/aminoacyl−tRNA synthetase pair for the in vivo incorporation of unnatural amino acids into proteins. J. Am. Chem. Soc. 2000, 122, 5010–5011. [Google Scholar] [CrossRef]
- Fechter, P.; Rudinger-Thirion, J.; Tukalo, M.; Giegé, R. Major tyrosine identity determinants in Methanococcus jannaschii and Saccharomyces cerevisiae tRNATyr are conserved but expressed differently. Eur. J. Biochem. 2001, 268, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Iraha, F.; Ohtake, K.; Sakamoto, K. Pyrrolysyl-tRNA synthetase with a unique architecture enhances the availability of lysine derivatives in synthetic genetic codes. Molecules 2018, 23, 2460. [Google Scholar] [CrossRef] [PubMed]
- Yokogawa, T.; Watanabe, Y.; Kumazawa, Y.; Ueda, T.; Hirao, I.; Miura, K.; Watanabe, K. A novel cloverleaf structure found in mammalian mitochondrial tRNASerUCN. Nucleic Acids Res. 1991, 19, 6101–6105. [Google Scholar] [CrossRef]
- Watanabe, Y.; Kawai, G.; Yokogawa, T.; Hayashi, N.; Kumazawa, Y.; Ueda, T.; Nishikawa, K.; Hirao, I.; Miura, K.; Watanabe, K. Higher-order structure of bovine mitochondrial tRNASerUGA: Chemical modification and computer modeling. Nucleic Acids Res. 1994, 22, 5378–5384. [Google Scholar] [CrossRef]
- Mustoe, A.M.; Liu, X.; Lin, P.J.; Al-Hashimi, H.A.; Fierke, C.A.; BrooksIII, C.L. A non-canonical secondary structure stabilizes mitochondrial tRNASerUCN by reducing the entropic cost of tertiary folding. J. Am. Chem. Soc. 2015, 137, 3592–3599. [Google Scholar] [CrossRef]
- McClain, W.H.; Foss, K.; Jenkins, R.A.; Schneider, J. Rapid determination of nucleotides that define tRNAGly acceptor identity. Proc. Natl. Acad. Sci. USA 1991, 88, 6147–6151. [Google Scholar] [CrossRef] [PubMed]
- Kleina, L.G.; Masson, J.M.; Normanly, J.; Abelson, J.; Miller, J.H. Construction of Escherichia coli amber suppressor tRNA genes II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. J. Mol. Biol. 1990, 213, 705–717. [Google Scholar] [CrossRef]
- Normanly, J.; Kleina, L.G.; Masson, J.M.; Abelson, J.; Miller, J.H. Construction of Escherichia coli amber suppressor tRNA genes III. Determination of tRNA specificity. J. Mol. Biol. 1990, 213, 719–726. [Google Scholar] [CrossRef]
- Schulman, L.H.; Pelka, H. In vitro conversion of a methionine to a glutamine-acceptor tRNA. Biochemistry 1985, 24, 7309–7314. [Google Scholar] [CrossRef]
- Rould, M.A.; Perona, J.J.; Söll, D.; Steitz, T.A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNAGln and ATP at 2.8 Å resolution. Science 1989, 246, 1135–1142. [Google Scholar] [CrossRef] [PubMed]
- Rould, M.A.; Perona, J.J.; Steitz, T.A. Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature 1991, 352, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Jahn, M.; Rogers, M.J.; Söll, D. Anticodon and acceptor stem nucleotides in tRNAGln are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature 1991, 352, 258–260. [Google Scholar] [CrossRef] [PubMed]
- Hayase, Y.; Jahn, M.; Rogers, M.J.; Sylvers, L.A.; Koizumi, M.; Inoue, H.; Ohtsuka, E.; Söll, D. Recognition of bases in Escherichia coli tRNAGln by glutaminyl-tRNA synthetase: A complete identity set. EMBO J. 1992, 11, 4159–4165. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nureki, O.; Ishitani, R.; Yaremchuk, A.; Tukalo, M.; Cusack, S.; Sakamoto, K.; Yokoyama, S. Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat. Struct. Biol. 2003, 10, 425–432. [Google Scholar] [CrossRef]
- Yaremchuk, A.; Kriklivyi, I.; Tukalo, M.; Cusack, S. Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 2002, 21, 3829–3840. [Google Scholar] [CrossRef]
- Théobald-Dietrich, A.; Frugier, M.; Giegé, R.; Rudinger-Thirion, J. Atypical archaeal tRNA pyrrolysine transcript behaves towards EF-Tu as a typical elongator tRNA. Nucleic Acids Res. 2004, 32, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Ambrogelly, A.; Gundllapalli, S.; Herring, S.; Polycarpo, C.; Frauer, C.; Söll, D. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc. Natl. Acad. Sci. USA 2007, 104, 3141–3146. [Google Scholar] [CrossRef] [PubMed]
- Gundllapalli, S.; Ambrogelly, A.; Umehara, T.; Li, D.; Polycarpo, C.; Söll, D. Misacylation of pyrrolysine tRNA in vitro and in vivo. FEBS Lett. 2008, 582, 3353–3358. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, R.; Prat, L.; Aerni, H.R.; Ling, J.; Merryman, C.; Glass, J.I.; Rinehart, J.; Söll, D. Transfer RNA misidentification scrambles sense codon recoding. Chembiochem 2013, 14, 1967–1972. [Google Scholar] [CrossRef] [PubMed]
- Thyer, R.; Ellington, A.D. The role of tRNA in establishing new genetic codes. Biochemistry 2018. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.C.W.; Chin, J.W. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat. Chem. 2018, 10, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Meineke, B.; Heimgärtner, J.; Lafranchi, L.; Elsässer, S.J. Methanomethylophilus alvus Mx1201 provides basis for mutual orthogonal pyrrolysyl tRNA/aminoacyl-tRNA synthetase pairs in mammalian cells. ACS Chem. Biol. 2018, 13, 3087–3096. [Google Scholar] [CrossRef]
- Serfling, R.; Lorenz, C.; Etzel, M.; Schicht, G.; Böttke, T.; Mörl, M.; Coin, I. Designer tRNAs for efficient incorporation of non-canonical amino acids by the pyrrolysine system in mammalian cells. Nucleic Acids Res. 2018, 46, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.; Bröcker, M.J.; Prat, L.; Ip, K.; Chirathivat, N.; Feiock, A.; Veszprémi, M.; Söll, D. A synthetic tRNA for EF-Tu mediated selenocysteine incorporation in vivo and in vitro. FEBS Lett. 2015, 589, 2194–2199. [Google Scholar] [CrossRef]
- Bullock, T.L.; Sherlin, L.D.; Perona, J.J. Tertiary core rearrangements in a tight binding transfer RNA aptamer. Nat. Struct. Biol. 2000, 7, 497–504. [Google Scholar]
- Ostrov, N.; Landon, M.; Guell, M.; Kuznetsov, G.; Teramoto, J.; Cervantes, N.; Zhou, M.; Singh, K.; Napolitano, M.G.; Moosburner, M.; et al. Design, synthesis, and testing toward a 57-codon genome. Science 2016, 353, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Auld, D.S.; Schimmel, P. Switching recognition of two tRNA synthetases with an amino acid swap in a designed peptide. Science 1995, 267, 1994–1996. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Xiao, H.; Schultz, P.G. Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2012, 109, 14841–14846. [Google Scholar] [CrossRef] [PubMed]
- McClain, W.H.; Foss, K. Changing the acceptor identity of a transfer RNA by altering nucleotides in a “variable pocket”. Science 1988, 241, 1804–1807. [Google Scholar] [CrossRef] [PubMed]
- Kiga, D.; Sakamoto, K.; Sato, S.; Hirao, I.; Yokoyama, S. Shifted positioning of the anticodon nucleotide residues of amber suppressor tRNA species by Escherichia coli arginyl-tRNA synthetase. Eur. J. Biochem. 2001, 268, 6207–6213. [Google Scholar] [CrossRef] [PubMed]



| Variant | Amber Suppression | Sequence 1 |
|---|---|---|
| MJR | Cm200 | CCGGCGGUAGUUCAGCCUGGUAGAACGGCGGACUCUAAAUCCGCAUGUCGCUGGUUCAAAUCCGGCCCGCCGGACCA |
| MJR1 | Cm200 | CCGGCGGUAGUUCAGCAGGGCAGAACGGCGGACUCUAAAUCCGCAUGGCGCUGGUUCAAAUCCGGCCCGCCGGACCA |
| Mm tRNAPyl | N. D. | GGAAACCUGAUCAUGUAGAUCGAAUGGACUCUAAAUCCGUUCAGCCGGGUUAGAUUCCCGGGGUUUCCGCCA |
| PYLY1 | Cm100 | CCGGCGGUGAUCAUGUAGAUCGAAUGGACUCUAAAUCCGUUCAGGCUGGUUAGAUUCCGGCCCGCCGGACCA |
| PYLY2 | Cm100 | CGAAACCUGAUCAUGUAGAUCGAAUGGACUCUAAAUCCGUUCAGCCGGGUUAGAUUCCCGGGGUUUCGACCA |
| PYLY1 (G1C72) | < Cm25 | GCGGCGGUGAUCAUGUAGAUCGAAUGGACUCUAAAUCCGUUCAGGCUGGUUAGAUUCCGGCCCGCCGGACACCA |
| PYLY1 (U1A72) | < Cm25 | UCGGCGGUGAUCAUGUAGAUCGAAUGGACUCUAAAUCCGUUCAGGCUGGUUAGAUUCCGGCCCGCCGGAAACCA |
| PYLY1 (G73) | <Cm25 | CCGGCGGUGAUCAUGUAGAUCGAAUGGACUCUAAAUCCGUUCAGGCUGGUUAGAUUCCGGCCCGCCGGGCCA |
| PYLY1 (U73) | <Cm25 | CCGGCGGUGAUCAUGUAGAUCGAAUGGACUCUAAAUCCGUUCAGGCUGGUUAGAUUCCGGCCCGCCGGUCCA |
| PYLY Dh | Cm50 | CGGGGGUGGAUCGAAUAGAUCACACGGACUCUAAAUUCGUGCAGGCGGGUGAAACUCCCGUACUCCCGACCA |
| PYLY CMa | Cm200 | CGGGGACGGUCCGGCGACCAGCGGGUCUCUAAAACCUGCCAGCGGGGUUCGACGCCCCGGUCUCUGACCA |
| Variant | Amber Suppression | Sequence in the Core 1 |
|---|---|---|
| GGCT2 | Cm100 | 8CGGCUAUGUAAGCCA26…44GUCGCUGGUUAGAUUCCGGC65 |
| GGCT3 | Cm100 | 8AGGCUAUGUAAGCCG26…44GUUGCUGGUUAGAUUCCGGC65 |
| GGCT4 | Cm100 | 8AGGCUAUGUAAGCCA26…44GUUGCUGGUUAGAUUCCGGC65 |
| GGCT5 | Cm50 | 8UGGCUAUGUAAGCCG26…44GUCGCUGGUUAGAUUCCGGC65 |
| GCCC2 | Cm100 | 8UGCCCAUGUAGGGCG26…44UGAGCUGGUUAGAUUCCGGC65 |
| GCCC3 | Cm100 | 8UGCCCAUGUAGGGCA26…44GGAGCUGGUUAGAUUCCGGC65 |
| GCCC4 | Cm100 | 8UGCCCAUGUAGGGCG26…44UGUGCUGGUUAGAUUCCGGC65 |
| GCCC5 | Cm100 | 8UGCCCAUGUAGGGCG26…44GGAGCUGGUUAGAUUCCGGC65 |
| GTTC1 | Cm100 | 8UGUUCAUGUAGAACG26…44UGAGCUGGUUAGAUUCCGGC65 |
| GTTC2 | Cm100 | 8UGUUCAUGUAGAACA26…44AGAGCUGGUUAGAUUCCGGC65 |
| GTTC3 | Cm100 | 8UGUUCAUGUAGAACA26…44CGAGCUGGUUAGAUUCCGGC65 |
| GTTC4 | Cm100 | 8AGUUCAUGUAGAACG26…44UGAGCUGGUUAGAUUCCGGC65 |
| GTTC5 | Cm100 | 8UGUUCAUGUAGAACG26…44AGAGCUGGUUAGAUUCCGGC65 |
| GTTC6 | Cm100 | 8UGUUCAUGUAGAACG26…44UGCGCUGGUUAGAUUCCGGC65 |
| GTTC7 | Cm100 | 8UGUUCAUGUAGAACA26…44UGAGCUGGUUAGAUUCCGGC65 |
| GTTC8 | Cm100 | 8UGUUCAUGUAGAACG26…44UGUGCUGGUUAGAUUCCGGC65 |
| Variant | Amber Suppression | Sequence in the Core 1 |
|---|---|---|
| DLE1 | Cm100 | 8UGAUCUUGUAGAUCG26…44UGAGCUGGUUAGAUUCCGGC65 |
| DLE2 | Cm100 | 8AGAUCAUGUUGAUCA26…44AGCGCUGGUUAGAUUCCGGC65 |
| DLE3 | Cm100 | 8UGAUCGUGUCGAUCG26…44AGAGCUGGUUAGAUUCCGGC65 |
| DLE4 | Cm200 | 8AGAUCCUGUGGAUCG26…44CUGGCUGGUUAGAUUCCGGC65 |
| DLE5 | Cm100 | 8UGAUCAUGUUGAUCC26…44UGCGCUGGUUAGAUUCCGGC65 |
| Variant | Amber Suppression | Sequence in the Core 1 |
|---|---|---|
| DT2-1 | Cm200 | 8UGAUCCUGGGGAUCG26…44CAGGCUGGUUCGAUUCCGGC65 |
| DT2-2 | Cm100 | 8UGAUCAAGGUGAUCG26…44CAGGCUGGUUCAAUUCCGGC65 |
| DT2-3 | Cm100 | 8UGAUCAUGGUGAUCG26…44CAGGCUGGUUCAACUCCGGC65 |
| DT2-4 | Cm50 | 8UGAUCAAGGAGAUCG26…44CAGGCUGGUUCGAUCCCGGC65 |
| DT3-1 | Cm200 | 8UGGCUCUGGGAGCCA26…44CUCGCUGGUUCGACUCCGGC65 |
| DT3-2 | Cm200 | 8UGGCUACGGUAGCCA26…44CUCGCUGGUUCGACCCCGGC65 |
| DT3-3 | Cm100 | 8UGGCUCUGGUAGCCA26…44CUCGCUGGUUCGAUUCCGGC65 |
| DT3-4 | Cm50 | 8UGGCUCGGGGAGCCA26…44CUCGCUGGUUCGAGUCCGGC65 |
| DT3-5 | Cm100 | 8UGGCUUAGGGAGCCA26…44CUCGCUGGUUCGAUUCCGGC65 |
| Variant | Amber Suppression | Sequence in the Core 1 |
|---|---|---|
| DT4-1 | Cm25 | 8AAGUUCAAGGUGAACG26…44AGGUGCUGGUUAGAUUCCGGC5 |
| DT4-2 | Cm50 | 8UAGUUCGUACUGAACG26…44AGUUGCUGGUUAGAUUCCGGC65 |
| DT5-1 | Cm100 | 8UAGUUCAACAAAGAACG26…44AAAUGCUGGUUAGAUUCCGGC65 |
| DT5-2 | Cm50 | 8UAGUUCAAAGGGGAACG26…44AAGUGCUGGUUAGAUUCCGGC65 |
| DT5-3 | Cm100 | 8UGGUUCAAAGUCGAACG26…44AGGAGCUGGUUAGAUUCCGGC65 |
| DT5-4 | Cm100 | 8UAGUUCAAACAAGAACG26…44UGGUGCUGGUUAGAUUCCGGC65 |
| DT5-5 | Cm200 | 8UAGUUCAACAUAGAACG26…44AGGUGCUGGUUAGAUUCCGGC65 |
| DT5-6 | Cm100 | 8UAGUUCAUAGUAGAACG26…44AGGUGCUGGUUAGAUUCCGGC65 |
| DT5-7 | Cm100 | 8UAGUUCAUGUCAGAACG26…44AAGAGCUGGUUAGAUUCCGGC65 |
| DT5-8 | Cm100 | 8AAGUUCAACGCGGAACG26…44AGGAGCUGGUUAGAUUCCGGC65 |
| DT5-9 | Cm100 | 8UAGUUCAAUGGAGAACG26…44CUGUGCUGGUUAGAUUCCGGC65 |
| DT5-10 | Cm100 | 8UGGUUCAGAGUAGAACG26…44AGAUGCUGGUUAGAUUCCGGC65 |
| DT5-11 | Cm100 | 8UGGUUCAAUUAAGAACG26…44AGGUGCUGGUUAGAUUCCGGC65 |
| DT6-1 | Cm100 | 8UAGUUCACUUGUAGAACG26…44AGGAGCUGGUUAGAUUCCGGC65 |
| DT6-2 | Cm100 | 8UAGUUCAACUUUAGAACG26…44GAAUGCUGGUUAGAUUCCGGC65 |
| Variant | Amber Suppression 1 | Sequence 2 |
|---|---|---|
| GLY1 | Cm15 | GCGGCGGUGGCUGUGGUAGCCAAAUGGACUCUAAAUCCGUUCUCGCUGGUUCGAGUCCGGCCCGCCGGACUCCA |
| GLY2 | Cm5 | GCGGCGGUGGCUGGGGUAGCCAAAUGGACUCUAAAUCCGUUCUCGCUGG UUCGAUUCCGGCCCGCCGGACUCCA |
| GLY3 | Cm5 | GCGGCGGUGGCUAUGGUAGCCAAAUGGACUCUAAAUCCGUUCUCGCUGG UUCGAGCCCGGCCCGCCGGACUCCA |
| GLY4 | Cm5 | GCGGCGGUGGCUCAGGAAGCCAAAUGGACUCUAAAUCCGUUCUCGCUGG UUCGAUUCCGGCCCGCCGGACUCCA |
| GLN1 | Cm15 | UCGGCGGUGGCUAUGGUAGCCAAAUGGACUCUAAAUCCGUUCUCGCUGG UUCGACUCCGGCCCGCCGGACGCCA |
| GLN2 | Cm5 | UCGGCGGUGGCUCUGGGAGCCAAAUGGACUCUAAAUCCGUUCUCGCUGG UUCGACUCCGGCCCGCCGGAUGCCA |
| GLNa | Cm15—25 | UGGGCGGUGGCUAUGGUAGCCAAAUGGACUCUAAAUCCGUUCUCGCUGG UUCGACUCCGGCCCGCCCAGCCA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakamoto, K.; Hayashi, A. Synthetic Tyrosine tRNA Molecules with Noncanonical Secondary Structures. Int. J. Mol. Sci. 2019, 20, 92. https://doi.org/10.3390/ijms20010092
Sakamoto K, Hayashi A. Synthetic Tyrosine tRNA Molecules with Noncanonical Secondary Structures. International Journal of Molecular Sciences. 2019; 20(1):92. https://doi.org/10.3390/ijms20010092
Chicago/Turabian StyleSakamoto, Kensaku, and Akiko Hayashi. 2019. "Synthetic Tyrosine tRNA Molecules with Noncanonical Secondary Structures" International Journal of Molecular Sciences 20, no. 1: 92. https://doi.org/10.3390/ijms20010092
APA StyleSakamoto, K., & Hayashi, A. (2019). Synthetic Tyrosine tRNA Molecules with Noncanonical Secondary Structures. International Journal of Molecular Sciences, 20(1), 92. https://doi.org/10.3390/ijms20010092