Surface Orientation and Binding Strength Modulate Shape of FtsZ on Lipid Surfaces
Abstract
:1. Introduction
2. Results
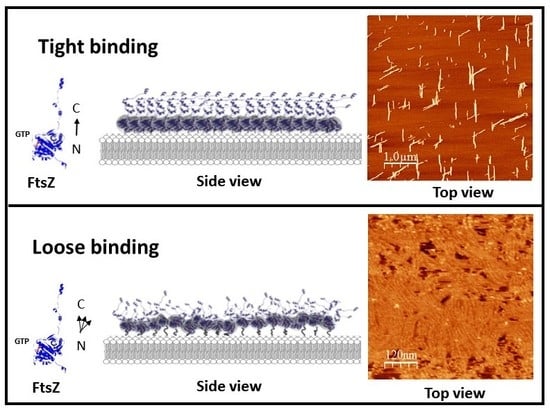
2.1. Tight Binding
2.2. Loose Binding
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Wang, X.; Lutkenhaus, J. Ftsz ring: The eubacterial division apparatus conserved in archaebacteria. Mol. Microbiol. 1996, 21, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Gilson, P.R.; Beech, P.L. Cell division protein ftsz: Running rings around bacteria, chloroplasts and mitochondria. Res. Microbiol. 2001, 152, 3–10. [Google Scholar] [CrossRef]
- Mingorance, J.; Rivas, G.; Vélez, M.; Gómez-Puertas, P.; Vicente, M. Strong ftsz is with the force: Mechanisms to constrict bacteria. Trends Microbiol. 2010, 18, 348–356. [Google Scholar] [CrossRef]
- Mateos-Gil, P.; Tarazona, P.; Vélez, M. Bacterial cell division: Modeling ftsz assembly and force generation from single filament experimental data. FEMS Microbiol. Rev. 2019, 43, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Yamane, J.; Mogi, N.; Yamaguchi, H.; Takemoto, H.; Yao, M.; Tanaka, I. Structural reorganization of the bacterial cell-division protein ftsz from staphylococcus aureus. Acta Crystall. Sect. D 2012, 68, 1175–1188. [Google Scholar] [CrossRef]
- Huecas, S.; Ramírez-Aportela, E.; Vergoñós, A.; Núñez-Ramírez, R.; Llorca, O.; Díaz, J.F.; Juan-Rodríguez, D.; Oliva, M.A.; Castellen, P.; Andreu, J.M. Self-organization of ftsz polymers in solution reveals spacer role of the disordered c-terminal tail. Biophys. J. 2017, 113, 1831–1844. [Google Scholar] [CrossRef]
- Nogales, E.; Wolf, S.G.; Downing, K.H. Structure of the αβ tubulin dimer by electron crystallography. Nature 1998, 391. [Google Scholar] [CrossRef]
- Oliva, M.A.; Cordell, S.C.; Lowe, J. Structural insights into ftsz protofilament formation. Nat. Struct. Mol. Biol. 2004, 11, 1243–1250. [Google Scholar] [CrossRef]
- Scheffers, D.J.; de Wit, J.G.; den Blaauwen, T.; Driessen, A.J. Gtp hydrolysis of cell division protein ftsz: Evidence that the active site is formed by the association of monomers. Biochemistry 2002, 41. [Google Scholar] [CrossRef]
- Martín-Galiano, A.J.; Buey, R.M.; Cabezas, M.; Andreu, J.M. Mapping flexibility and the assembly switch of cell division protein ftsz by computational and mutational approaches. J. Biol. Chem. 2010, 285, 22554–22565. [Google Scholar] [CrossRef]
- Wagstaff, J.M.; Tsim, M.; Oliva, M.A.; García-Sanchez, A.; Kureisaite-Ciziene, D.; Andreu, J.M.; Löwe, J. A polymerization-associated structural switch in ftsz that enables treadmilling of model filaments. mBio 2017, 8. [Google Scholar] [CrossRef]
- Buske, P.J.; Levin, P.A. A flexible c-terminal linker is required for proper ftsz assembly in vitro and cytokinetic ring formation in vivo. Mol. Microbiol. 2013, 89, 249–263. [Google Scholar] [CrossRef]
- Hernández-Rocamora, V.M.; Reija, B.; García, C.; Natale, P.; Alfonso, C.; Minton, A.P.; Zorrilla, S.; Rivas, G.; Vicente, M. Dynamic interaction of the escherichia coli cell division zipa and ftsz proteins evidenced in nanodiscs. J. Biol. Chem. 2012, 287, 30097–30104. [Google Scholar] [CrossRef]
- Márquez, I.F.; Mateos-Gil, P.; Shin, J.Y.; Lagos, R.; Monasterio, O.; Vélez, M. Mutations on ftsz lateral helix h3 that disrupt cell viability hamper reorganization of polymers on lipid surfaces. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1815–1827. [Google Scholar] [CrossRef]
- Pichoff, S.; Lutkenhaus, J. Tethering the z ring to the membrane through a conserved membrane targeting sequence in ftsa. Mol. Microbiol. 2005, 55, 1722–1734. [Google Scholar] [CrossRef]
- Hale, C.A.; de Boer, P.A.J. Direct binding of ftsz to zipa, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 1997, 88, 175–185. [Google Scholar] [CrossRef]
- Ohashi, T.; Hale, C.A.; de Boer, P.A.J.; Erickson, H.P. Structural evidence that the p/q domain of zipa is an unstructured, flexible tether between the membrane and the c-terminal ftsz-binding domain. J. Bacteriol. 2002, 184, 4313–4315. [Google Scholar] [CrossRef]
- Buske, P.J.; Mittal, A.; Pappu, R.V.; Levin, P.A. An intrinsically disordered linker plays a critical role in bacterial cell division. Semin. Cell Dev. Biol. 2015, 37, 3–10. [Google Scholar] [CrossRef]
- Sundararajan, K.; Miguel, A.; Desmarais, S.M.; Meier, E.L.; Casey Huang, K.; Goley, E.D. The bacterial tubulin ftsz requires its intrinsically disordered linker to direct robust cell wall construction. Nat. Commun. 2015, 6, 7281. [Google Scholar] [CrossRef]
- Sundararajan, K.; Goley, E.D. The intrinsically disordered c-terminal linker of ftsz regulates protofilament dynamics and superstructure in vitro. J. Biol. Chem. 2017, 292, 20509–20527. [Google Scholar] [CrossRef]
- Mateos-Gil, P.; Marquez, I.; Lopez-Navajas, P.; Jimenez, M.; Vicente, M.; Mingorance, J.; Rivas, G.; Velez, M. Ftsz polymers bound to lipid bilayers through zipa form dynamic two dimensional networks. Biochim. Biophys. Acta 2012, 1818, 806–813. [Google Scholar] [CrossRef]
- Encinar, M.; Kralicek, A.V.; Martos, A.; Krupka, M.; Cid, S.; Alonso, A.; Rico, A.I.; Jiménez, M.; Vélez, M. Polymorphism of ftsz filaments on lipid surfaces: Role of monomer orientation. Langmuir 2013, 29, 9436–9446. [Google Scholar] [CrossRef] [PubMed]
- Loose, M.; Mitchison, T.J. The bacterial cell division proteins ftsa and ftsz self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 2013, 16, 38–46. [Google Scholar] [CrossRef]
- Ramirez-Diaz, D.A.; García-Soriano, D.A.; Raso, A.; Mücksch, J.; Feingold, M.; Rivas, G.; Schwille, P. Treadmilling analysis reveals new insights into dynamic ftsz ring architecture. PLoS Biol. 2018, 16, e2004845. [Google Scholar] [CrossRef]
- Sundararajan, K.; Vecchiarelli, A.; Mizuuchi, K.; Goley, E.D. Species- and c-terminal linker-dependent variations in the dynamic behavior of ftsz on membranes in vitro. Mol. Microbiol. 2018, 110, 47–63. [Google Scholar] [CrossRef]
- Milam, S.L.; Osawa, M.; Erickson, H. Negative-stain electron microscopy of inside-out ftsz rings reconstituted on artificial membrane tubules show ribbons of protofilaments. Biophys. J. 2012, 103, 59–68. [Google Scholar] [CrossRef]
- Osawa, M.; Anderson, D.E.; Erickson, H.P. Reconstitution of contractile ftsz rings in liposomes. Science 2008, 320, 792–794. [Google Scholar] [CrossRef]
- Osawa, M.; Anderson, D.E.; Erickson, H.P. Curved ftsz protofilaments generate bending forces on liposome membranes. EMBO J. 2009, 28, 3476–3484. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez de Prado Salas, P.; Encinar, M.; Velez, M.; Tarazona, P. Ftsz protein on bilayer membranes: Effects of specific lateral bonds. Soft Matter 2013, 9, 6072–6079. [Google Scholar] [CrossRef]
- Paez, A.; Mateos-Gil, P.; Hörger, I.; Mingorance, J.; Rivas, G.; Vicente, M.; Vélez, M.; Tarazona, P. Simple modeling of ftsz polymers on flat and curved surfaces: Correlation with experimental in vitro observations. PMC Biophys. 2009, 2, 8. [Google Scholar] [CrossRef]
- Hörger, I.; Velasco, E.; Mingorance, J.; Rivas, G.; Tarazona, P.; Vélez, M. Langevin computer simulations of bacterial protein filaments and the force-generating mechanism during cell division. Phys. Rev. E 2008, 77, 011902. [Google Scholar] [CrossRef] [PubMed]
- Hörger, I.; Velasco, E.; Rivas, G.; Vélez, M.; Tarazona, P. Ftsz bacterial cytoskeletal polymers on curved surfaces: The importance of lateral interactions. Biophys. J. 2008, 94, L81–L83. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez de Prado Salas, P.; Horger, I.; Martin-Garcia, F.; Mendieta, J.; Alonso, A.; Encinar, M.; Gomez-Puertas, P.; Velez, M.; Tarazona, P. Torsion and curvature of ftsz filaments. Soft Matter 2014, 10, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- de Prado Salas, P.G.; Encinar, M.; Alonso, A.; Vélez, M.; Tarazona, P. Modeling the interplay between protein and lipid aggregation in supported membranes. Chem. Phys. Lipids 2015, 185, 141–152. [Google Scholar] [CrossRef] [PubMed]
- González de Prado Salas, P.; Tarazona, P. Collective effects of torsion in ftsz filaments. Phys. Rev. E 2016, 93, 042407. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, S.; Chwastek, G.; Fischer-Friedrich, E.; Ehrig, C.; Mönch, I.; Schwille, P. Surface topology engineering of membranes for the mechanical investigation of the tubulin homologue ftsz. Angew. Chem. Int. Ed. 2012, 51, 11858–11862. [Google Scholar] [CrossRef]
- Hsin, J.; Gopinathan, A.; Huang, K.C. Nucleotide-dependent conformations of ftsz dimers and force generation observed through molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 2012, 109, 9432–9437. [Google Scholar] [CrossRef]
- Qing-Ming, Y.; Lutkenhaus, J. The nucleotide sequence of the essential cell-division gene ftsz of escherichia coli. Gene 1985, 36, 241–247. [Google Scholar] [CrossRef]
- Nogales, E.; Downing, K.H.; Amos, L.A.; Löwe, J. Tubulin and ftsz form a distinct family of gtpases. Nat. Struct. Biol. 1998, 5. [Google Scholar] [CrossRef]
- Shahinian, S.; Silvius, J.R. A novel strategy affords high-yield coupling of antibody fab´fragments to liposomes. Biochim. Biophys. Acta Biomembr. 1995, 1239, 157–167. [Google Scholar] [CrossRef]
- Huwyler, J.; Wu, D.; Pardridge, W.M. Brain drug delivery of small molecules using immunoliposomes. Proc. Natl. Acad. Sci. USA 1996, 93, 14164–14169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mingorance, J.; Tadros, M.; Vicente, M.; Gonzalez, J.M.; Rivas, G.; Velez, M. Visualization of single escherichia coli ftsz filament dynamics with atomic force microscopy. J. Biol.Chem. 2005, 280, 20909–20914. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Erickson, H.P. Rapid in vitro assembly dynamics and subunit turnover of ftsz demonstrated by fluorescence resonance energy transfer. J. Biol. Chem. 2005, 280, 22549–22554. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Stricker, J.; Erickson, H.P. Site-specific mutations of ftsz - effects on gtpase and in vitro assembly. BMC Microbiol. 2001, 1, 7. [Google Scholar] [CrossRef]
- Shin, J.Y.; Vollmer, W.; Lagos, R.; Monasterio, O. Glutamate 83 and arginine 85 of helix h3 bend are key residues for ftsz polymerization, gtpase activity and cellular viability of escherichia coli: Lateral mutations affect ftsz polymerization and E. coli viability. BMC Microbiol. 2013, 13, 26. [Google Scholar] [CrossRef]
- Jaiswal, R.; Patel, R.Y.; Asthana, J.; Jindal, B.; Balaji, P.V.; Panda, D. E93r substitution of escherichia coli ftsz induces bundling of protofilaments, reduces gtpase activity, and impairs bacterial cytokinesis. J. Biol. Chem. 2010, 285, 31796–31805. [Google Scholar] [CrossRef] [PubMed]
- Monahan, L.G.; Robinson, A.; Harry, J.E. Lateral ftsz association and the assembly of the cytokinetic z ring in bacteria. Mol. Microbiol. 2009, 74. [Google Scholar] [CrossRef] [PubMed]
- Guan, F.; Yu, J.; Yu, J.; Liu, Y.; Li, Y.; Feng, X.-H.; Huang, K.C.; Chang, Z.; Ye, S. Lateral interactions between protofilaments of the bacterial tubulin homolog ftsz are essential for cell division. eLife 2018, 7, e35578. [Google Scholar] [CrossRef]
- Fontaine, S.D.; Reid, R.; Robinson, L.; Ashley, G.W.; Santi, D.V. Long-term stabilization of maleimide–thiol conjugates. Bioconj. Chem. 2015, 26, 145–152. [Google Scholar] [CrossRef]
- Watkins, E.B.; El-khouri, R.J.; Miller, C.E.; Seaby, B.G.; Majewski, J.; Marques, C.M.; Kuhl, T.L. Structure and thermodynamics of lipid bilayers on polyethylene glycol cushions: Fact and fiction of peg cushioned membranes. Langmuir 2011, 27, 13618–13628. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, Y.; Zhang, F.; Pillai, S.; Liu, J.; Li, R.; Dai, B.; Li, B.; Zhang, Y. Hierarchical ordering of amyloid fibrils on the mica surface. Nanoscale 2013, 5, 4816–4822. [Google Scholar] [CrossRef] [PubMed]
- Bisson-Filho, A.W.; Hsu, Y.-P.; Squyres, G.R.; Kuru, E.; Wu, F.; Jukes, C.; Sun, Y.; Dekker, C.; Holden, S.; VanNieuwenhze, M.S.; et al. Treadmilling by ftsz filaments drives peptidoglycan synthesis and bacterial cell division. Science 2017, 355, 739. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lyu, Z.; Miguel, A.; McQuillen, R.; Huang, K.C.; Xiao, J. Gtpase activity–coupled treadmilling of the bacterial tubulin ftsz organizes septal cell wall synthesis. Science 2017, 355, 744. [Google Scholar] [CrossRef] [PubMed]
- Torrance, L.; Ziegler, A.; Pittman, H.; Paterson, M.; Toth, R.; Eggleston, I. Oriented immobilisation of engineered single-chain antibodies to develop biosensors for virus detection. J. Virol. Methods 2006, 134, 164–170. [Google Scholar] [CrossRef]
- Rivas, G.; López, A.; Mingorance, J.; Ferrándiz, M.a.J.; Zorrilla, S.; Minton, A.P.; Vicente, M.; Andreu, J.M. Magnesium-induced linear self-association of the ftsz bacterial cell division protein monomer: The primary steps for ftsz assembly. J. Biol. Chem. 2000, 275, 11740–11749. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Herrero, F.; Pablo, P.J.D.; Fernández-Sánchez, R.; Colchero, J.; Gómez-Herrero, J.; Baró, A.M. Scanning force microscopy jumping and tapping modes in liquids. Appl. Phys. Lett. 2002, 81, 2620–2622. [Google Scholar] [CrossRef] [Green Version]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Márquez, I.; Díaz-Haro, G.; Vélez, M. Surface Orientation and Binding Strength Modulate Shape of FtsZ on Lipid Surfaces. Int. J. Mol. Sci. 2019, 20, 2545. https://doi.org/10.3390/ijms20102545
Márquez I, Díaz-Haro G, Vélez M. Surface Orientation and Binding Strength Modulate Shape of FtsZ on Lipid Surfaces. International Journal of Molecular Sciences. 2019; 20(10):2545. https://doi.org/10.3390/ijms20102545
Chicago/Turabian StyleMárquez, Ileana, Gabriel Díaz-Haro, and Marisela Vélez. 2019. "Surface Orientation and Binding Strength Modulate Shape of FtsZ on Lipid Surfaces" International Journal of Molecular Sciences 20, no. 10: 2545. https://doi.org/10.3390/ijms20102545
APA StyleMárquez, I., Díaz-Haro, G., & Vélez, M. (2019). Surface Orientation and Binding Strength Modulate Shape of FtsZ on Lipid Surfaces. International Journal of Molecular Sciences, 20(10), 2545. https://doi.org/10.3390/ijms20102545