Distribution and Diversity of Cytochrome P450 Monooxygenases in the Fungal Class Tremellomycetes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Pathogenic Cryptococcal Species Have Few CYPs in Their Genomes
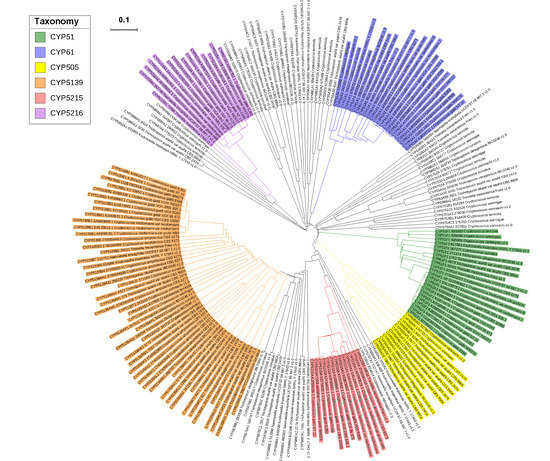
2.2. New CYP Families Were Found in Tremellomycetes
2.3. Four CYP Families Are Conserved in Pathogenic Cryptococcal Species
2.4. Pathogenic Cryptococcal Species Have the Highest CYP Diversity
2.5. Most of the CYPs from the Species of Tremellomycetes Are Orphans with No Known Function
3. Materials and Methods
3.1. Species and Databases
3.2. CYP Mining and Annotation
3.3. Phylogenetic Analysis of CYPs
3.4. Generation of CYP Profile Heat-Maps
3.5. CYP Diversity Percentage Analysis
3.6. Functional Prediction of CYPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- May, R.C.; Stone, N.R.; Wiesner, D.L.; Bicanic, T.; Nielsen, K. Cryptococcus: from environmental saprophyte to global pathogen. Nat. Rev. Microbiol. 2016, 14, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Loftus, B.J.; Fung, E.; Roncaglia, P.; Rowley, D.; Amedeo, P.; Bruno, D.; Vamathevan, J.; Miranda, M.; Anderson, I.J.; Fraser, J.A.; et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 2005, 307, 1321–1324. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, C.A.; Kronstad, J.W.; Taylor, G.; Warren, R.; Yuen, M.; Hu, G.; Jung, W.H.; Sham, A.; Kidd, S.E.; Tangen, K.; et al. Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. MBio 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Farrer, R.A.; Desjardins, C.A.; Sakthikumar, S.; Gujja, S.; Saif, S.; Zeng, Q.; Chen, Y.; Voelz, K.; Heitman, J.; May, R.C.; et al. Genome evolution and innovation across the four major lineages of Cryptococcus gattii. MBio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Cryptococcal Disease: What’s New and Important. Available online: https://www.who.int/hiv/mediacentre/news/cryptococcal-disease-key-messages/en/ (accessed on 8 April 2019).
- Rajasingham, R.; Smith, R.M.; Park, B.J.; Jarvis, J.N.; Govender, N.P.; Chiller, T.M.; Denning, D.W.; Loyse, A.; Boulware, D.R. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017, 17, 873–881. [Google Scholar] [CrossRef]
- Passer, A.R.; Coelho, M.A.; Billmyre, R.B.; Nowrousian, M.; Mittelbach, M.; Yurkov, A.M.; Averette, A.F.; Cuomo, C.A.; Sun, S.; Heitman, J. Genetic and genomic analyses reveal boundaries between species closely related to Cryptococcus pathogens. bioRxiv 2019. [Google Scholar] [CrossRef]
- Kuo, A.; Salamov, A.; Korzeniewski, F.; Nordberg, H.; Shabalov, I.; Dubchak, I.; Otillar, R.; Riley, R.; Ohm, R.; Nikitin, R.; et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2013, 42, D699–D704. [Google Scholar]
- Close, D.; Ojumu, J.; Zhang, G. Draft genome sequence of Cryptococcus terricola JCM 24523, an oleaginous yeast capable of expressing exogenous DNA. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Tanimura, A.; Takashima, M.; Sugita, T.; Endoh, R.; Kikukawa, M.; Yamaguchi, S.; Sakuradani, E.; Ogawa, J.; Ohkuma, M.; Shima, J. Cryptococcus terricola is a promising oleaginous yeast for biodiesel production from starch through consolidated bioprocessing. Sci. Rep. 2014, 4, 4776. [Google Scholar] [CrossRef]
- Close, D.; Ojumu, J. Draft Genome sequence of the oleaginous yeast Cryptococcus curvatus ATCC 20509. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Nowicka, J.; Nawrot, U.; Haus, O.; Kuliczkowski, K.; Fonteyne, P.-A.; Nolard, N. Cryptococcus curvatus in peritoneal fluid of gastric lymphoma patient with complex chromosome aberrations—Case report. Med. Mycol. Mikol. 2007, 14, 285–287. [Google Scholar]
- Vishniac, H.S.; Hempfling, W.P. Cryptococcus vishniacii sp. nov. an Antarctic yeast. Int. J. Syst. Evol. Microbiol. 1979, 29, 153–158. [Google Scholar] [CrossRef]
- Schmidt, S.K.; Vimercati, L.; Darcy, J.L.; Arán, P.; Gendron, E.M.; Solon, A.J.; Porazinska, D.; Dorador, C. A Naganishia in high places: functioning populations or dormant cells from the atmosphere? Mycology 2017, 8, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Milanović, V.; Comitini, F.; Ciani, M. Grape berry yeast communities: influence of fungicide treatments. Int. J. Food Microbiol. 2013, 161, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Yadav, V.; Billmyre, R.B.; Cuomo, C.A.; Nowrousian, M.; Wang, L.; Souciet, J.L.; Boekhout, T.; Porcel, B.; Wincker, P.; et al. Fungal genome and mating system transitions facilitated by chromosomal translocations involving intercentromeric recombination. PLoS Biol. 2017, 15, e2002527. [Google Scholar] [CrossRef] [PubMed]
- Mondo, S.J.; Dannebaum, R.O.; Kuo, R.C.; Louie, K.B.; Bewick, A.J.; LaButti, K.; Haridas, S.; Kuo, A.; Salamov, A.; Ahrendt, S.R.; et al. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 2017, 49, 964–968. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.E.; Thornton, C.R. Differentiation of the emerging human pathogens Trichosporon asahii and Trichosporon asteroides from other pathogenic yeasts and moulds by using species-specific monoclonal antibodies. PLoS ONE 2014, 9, e84789. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Ao, J.; Wang, W.; Song, K.; Li, R.; Wang, D. Disseminated trichosporonosis in China. Mycoses 2003, 46, 519–523. [Google Scholar] [CrossRef]
- Yang, R.Y.; Li, H.T.; Zhu, H.; Zhou, G.P.; Wang, M.; Wang, L. Genome sequence of the Trichosporon asahii environmental strain CBS 8904. Eukaryot. Cell 2012, 11, 1586–1587. [Google Scholar] [CrossRef]
- Kourist, R.; Bracharz, F.; Lorenzen, J.; Kracht, O.N.; Chovatia, M.; Daum, C.; Deshpande, S.; Lipzen, A.; Nolan, M.; Ohm, R.A.; et al. Genomics and transcriptomics analyses of the oil-accumulating basidiomycete yeast Trichosporon oleaginosus: Insights into substrate utilization and alternative evolutionary trajectories of fungal mating systems. MBio 2015, 6, e00918. [Google Scholar] [CrossRef]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martínez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.S. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef] [PubMed]
- Perfect, J.R.; Dismukes, W.E.; Dromer, F.; Goldman, D.L.; Graybill, J.R.; Hamill, R.J.; Harrison, T.S.; Larsen, R.A.; Lortholary, O.; Nguyen, M.H.; et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clin. Infect. Dis. 2010, 50, 291–322. [Google Scholar] [CrossRef] [PubMed]
- Hanson, K.E.; Catania, J.; Alexander, B.D.; Perfect, J.R. Drug Resistance in Cryptococcosis. In Antimicrobial Drug Resistance: Clinical and Epidemiological Aspects; Mayers, D.L., Sobel, J.D., Ouellette, M., Kaye, K.S., Marchaim, D., Eds.; Springer International Publishing: Cham, Switzerland, 2017; Volume 2, pp. 1119–1140. [Google Scholar]
- Kelly, S.L.; Kelly, D.E. Microbial cytochromes P450: biodiversity and biotechnology. Where do cytochromes P450 come from, what do they do and what can they do for us? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368. [Google Scholar] [CrossRef] [PubMed]
- Lamb, D.C.; Corran, A.; Baldwin, B.C.; Kwon-Chung, J.; Kelly, S.L. Resistant P45051A1 activity in azole antifungal tolerant Cryptococcus neoformans from AIDS patients. FEBS Lett. 1995, 368, 326–330. [Google Scholar] [CrossRef]
- Sionov, E.; Chang, Y.C.; Garraffo, H.M.; Dolan, M.A.; Ghannoum, M.A.; Kwon-Chung, K.J. Identification of a Cryptococcus neoformans cytochrome P450 lanosterol 14alpha-demethylase (Erg11) residue critical for differential susceptibility between fluconazole/voriconazole and itraconazole/posaconazole. Antimicrob. Agents Chemother. 2012, 56, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- Rodero, L.; Mellado, E.; Rodriguez, A.C.; Salve, A.; Guelfand, L.; Cahn, P.; Cuenca-Estrella, M.; Davel, G.; Rodriguez-Tudela, J.L. G484S amino acid substitution in lanosterol 14-alpha demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob. Agents Chemother. 2003, 47, 3653–3656. [Google Scholar] [CrossRef] [PubMed]
- Ngamskulrungroj, P.; Chang, Y.; Hansen, B.; Bugge, C.; Fischer, E.; Kwon-Chung, K.J. Characterization of the chromosome 4 genes that affect fluconazole-induced disomy formation in Cryptococcus neoformans. PLoS ONE 2012, 7, e33022. [Google Scholar] [CrossRef]
- Revankar, S.G.; Fu, J.; Rinaldi, M.G.; Kelly, S.L.; Kelly, D.E.; Lamb, D.C.; Keller, S.M.; Wickes, B.L. Cloning and characterization of the lanosterol 14alpha-demethylase (ERG11) gene in Cryptococcus neoformans. Biochem. Biophys. Res. Commun. 2004, 324, 719–728. [Google Scholar] [CrossRef]
- Kushima, H.; Tokimatsu, I.; Ishii, H.; Kawano, R.; Shirai, R.; Kishi, K.; Hiramatsu, K.; Kadota, J. Cloning of the lanosterol 14-alpha-demethylase ( ERG11) gene in Trichosporon asahii: A possible association between G453R amino acid substitution and azole resistance in T. asahii. FEMS Yeast Res. 2012, 12, 662–667. [Google Scholar] [CrossRef]
- Kushima, H.; Tokimatsu, I.; Ishii, H.; Kawano, R.; Watanabe, K.; Kadota, J.I. A new amino acid substitution at G150S in lanosterol 14-alpha eemethylase (Erg11 protein) in multi-azole-resistant Trichosporon asahii. Med. Mycol. J. 2017, 58, E23–E28. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Fothergill, A.W.; Iqbal, N.; Bolden, C.B.; Grossman, N.T.; Garvey, E.P.; Brand, S.R.; Hoekstra, W.J.; Schotzinger, R.J.; Ottinger, E.; et al. The investigational fungal Cyp51 inhibitor VT-1129 demonstrates potent in vitro activity against Cryptococcus neoformans and Cryptococcus gattii. Antimicrob. Agents Chemother. 2016, 60, 2528–2531. [Google Scholar] [CrossRef] [PubMed]
- Warrilow, A.G.; Parker, J.E.; Price, C.L.; Nes, W.D.; Garvey, E.P.; Hoekstra, W.J.; Schotzinger, R.J.; Kelly, D.E.; Kelly, S.L. The investigational drug VT-1129 is a highly potent inhibitor of Cryptococcus species CYP51 but only weakly inhibits the human enzyme. Antimicrob. Agents Chemother. 2016, 60, 4530–4538. [Google Scholar] [CrossRef]
- Nielsen, K.; Vedula, P.; Smith, K.D.; Meya, D.B.; Garvey, E.P.; Hoekstra, W.J.; Schotzinger, R.J.; Boulware, D.R. Activity of VT-1129 against Cryptococcus neoformans clinical isolates with high fluconazole MICs. Med. Mycol. 2017, 55, 453–456. [Google Scholar] [PubMed]
- Qhanya, L.B.; Matowane, G.; Chen, W.; Sun, Y.; Letsimo, E.M.; Parvez, M.; Yu, J.H.; Mashele, S.S.; Syed, K. Genome-wide annotation and comparative analysis of cytochrome P450 monooxygenases in Basidiomycete biotrophic plant pathogens. PLoS ONE 2015, 10, e0142100. [Google Scholar] [CrossRef] [PubMed]
- Syed, K.; Shale, K.; Pagadala, N.S.; Tuszynski, J. Systematic identification and evolutionary analysis of catalytically versatile cytochrome P450 monooxygenase families enriched in model basidiomycete fungi. PLoS ONE 2014, 9, e86683. [Google Scholar] [CrossRef] [PubMed]
- Kgosiemang, I.K.R.; Syed, K.; Mashele, S.S. Comparative genomics and evolutionary analysis of cytochrome P450 monooxygenases in fungal subphylum Saccharomycotina. J. Pure Appl. Microbiol. 2014, 8, 291–302. [Google Scholar]
- Duarte-Oliveira, C.; Rodrigues, F.; Goncalves, S.M.; Goldman, G.H.; Carvalho, A.; Cunha, C. The cell biology of the Trichosporon-host interaction. Front. Cell. Infect. Microbiol. 2017, 7, 118. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 nomenclature. Methods Mol. Biol. 1998, 107, 15–24. [Google Scholar] [PubMed]
- Nelson, D.R. Cytochrome P450 nomenclature, 2004. Methods Mol. Biol. 2006, 320, 1–10. [Google Scholar] [PubMed]
- Nelson, D.R. The cytochrome p450 homepage. Hum. Genom. 2009, 4, 59–65. [Google Scholar]
- Kelly, S.L.; Lamb, D.C.; Baldwin, B.C.; Corran, A.J.; Kelly, D.E. Characterization of Saccharomyces cerevisiae CYP61, sterol delta22-desaturase, and inhibition by azole antifungal agents. J. Biol. Chem. 1997, 272, 9986–9988. [Google Scholar] [CrossRef]
- Nakayama, N.; Takemae, A.; Shoun, H. Cytochrome P450foxy, a catalytically self-sufficient fatty acid hydroxylase of the fungus Fusarium oxysporum. J. Biochem. 1996, 119, 435–440. [Google Scholar] [CrossRef]
- Mingot, J.M.; Penalva, M.A.; Fernandez-Canon, J.M. Disruption of phacA, an Aspergillus nidulans gene encoding a novel cytochrome P450 monooxygenase catalyzing phenylacetate 2-hydroxylation, results in penicillin overproduction. J. Biol. Chem. 1999, 274, 14545–14550. [Google Scholar] [CrossRef]
- Faber, B.W.; van Gorcom, R.F.; Duine, J.A. Purification and characterization of benzoate-para-hydroxylase, a cytochrome P450 (CYP53A1), from Aspergillus niger. Arch. Biochem. Biophys. 2001, 394, 245–254. [Google Scholar] [CrossRef]
- Matsuzaki, F.; Wariishi, H. Molecular characterization of cytochrome P450 catalyzing hydroxylation of benzoates from the white-rot fungus Phanerochaete chrysosporium. Biochem. Biophys. Res. Commun. 2005, 334, 1184–1190. [Google Scholar] [CrossRef]
- Durairaj, P.; Jung, E.; Park, H.H.; Kim, B.G.; Yun, H. Comparative functional characterization of a novel benzoate hydroxylase cytochrome P450 of Fusarium oxysporum. Enzym. Microb. Technol. 2015, 70, 58–65. [Google Scholar] [CrossRef]
- Jawallapersand, P.; Mashele, S.S.; Kovacic, L.; Stojan, J.; Komel, R.; Pakala, S.B.; Krasevec, N.; Syed, K. Cytochrome P450 monooxygenase CYP53 family in fungi: Comparative structural and evolutionary analysis and its role as a common alternative anti-fungal drug target. PLoS ONE 2014, 9, e107209. [Google Scholar] [CrossRef]
- Nakahara, K.; Tanimoto, T.; Hatano, K.; Usuda, K.; Shoun, H. Cytochrome P-450 55A1 (P-450dNIR) acts as nitric oxide reductase employing NADH as the direct electron donor. J. Biol. Chem. 1993, 268, 8350–8355. [Google Scholar]
- Shoun, H.; Fushinobu, S.; Jiang, L.; Kim, S.W.; Wakagi, T. Fungal denitrification and nitric oxide reductase cytochrome P450nor. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 1186–1194. [Google Scholar] [CrossRef] [Green Version]
- Basenko, E.Y.; Pulman, J.A.; Shanmugasundram, A.; Harb, O.S.; Crouch, K.; Starns, D.; Warrenfeltz, S.; Aurrecoechea, C.; Stoeckert, C.J., Jr.; Kissinger, J.C.; et al. FungiDB: An integrated bioinformatic resource for fungi and Oomycetes. J. Fungi 2018, 4, 39. [Google Scholar] [CrossRef]
- Matowane, R.G.; Wieteska, L.; Bamal, H.D.; Kgosiemang, I.K.R.; Van Wyk, M.; Manume, N.A.; Abdalla, S.M.H.; Mashele, S.S.; Gront, D.; Syed, K. In silico analysis of cytochrome P450 monooxygenases in chronic granulomatous infectious fungus Sporothrix schenckii: Special focus on CYP51. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 166–177. [Google Scholar] [CrossRef]
- Ngwenya, M.L.; Chen, W.; Basson, A.K.; Shandu, J.S.; Yu, J.H.; Nelson, D.R.; Syed, K. Blooming of unusual cytochrome P450s by tandem duplication in the pathogenic fungus Conidiobolus coronatus. Int. J. Mol. Sci. 2018, 19, 1711. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Bamal, H.D.; Chen, W.; Mashele, S.S.; Nelson, D.R.; Kappo, A.P.; Mosa, R.A.; Yu, J.H.; Tuszynski, J.A.; Syed, K. Comparative analyses and structural insights of the novel cytochrome P450 fusion protein family CYP5619 in Oomycetes. Sci. Rep. 2018, 8, 6597. [Google Scholar] [CrossRef] [Green Version]
- Senate, L.M.; Tjatji, M.P.; Pillay, K.; Chen, W.; Zondo, N.M.; Syed, P.R.; Mnguni, F.C.; Chiliza, Z.E.; Bamal, H.D.; Karpoormath, R.; et al. Similarities, variations, and evolution of cytochrome P450s in Streptomyces versus Mycobacterium. Sci. Rep. 2019, 9, 3962. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Mthethwa, B.C.; Chen, W.; Ngwenya, M.L.; Kappo, A.P.; Syed, P.R.; Karpoormath, R.; Yu, J.H.; Nelson, D.R.; Syed, K. Comparative analyses of cytochrome P450s and those associated with secondary metabolism in Bacillus species. Int. J. Mol. Sci. 2018, 19, 3623. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. BioTechniques 2003, 34, 374–378. [Google Scholar] [CrossRef]
- Parvez, M.; Qhanya, L.B.; Mthakathi, N.T.; Kgosiemang, I.K.; Bamal, H.D.; Pagadala, N.S.; Xie, T.; Yang, H.; Chen, H.; Theron, C.W.; et al. Molecular evolutionary dynamics of cytochrome P450 monooxygenases across kingdoms: Special focus on mycobacterial P450s. Sci. Rep. 2016, 6, 33099. [Google Scholar] [CrossRef]
- Sello, M.M.; Jafta, N.; Nelson, D.R.; Chen, W.; Yu, J.H.; Parvez, M.; Kgosiemang, I.K.; Monyaki, R.; Raselemane, S.C.; Qhanya, L.B.; et al. Diversity and evolution of cytochrome P450 monooxygenases in Oomycetes. Sci. Rep. 2015, 5, 11572. [Google Scholar] [CrossRef]





| Species Name | Information | References |
|---|---|---|
| Cryptococcus neoformans | C. neoformans causes meningitis in immunocompromised and apparently in immunocompetent humans. This organism is considered a major opportunistic pathogen and a leading cause of mortality in patients infected with HIV. | [2] |
| Cryptococcus gattii | C. gattii causes respiratory (pneumonia) and neurological (meningoencephalitis) diseases in humans and animals and it can infect immunocompetent hosts. | [3,4] |
| Cryptococcus terricola JCM 24523 | C. terricola is oleaginous yeast and has been suggested as a candidate for the consolidated bioprocessing of hydrocarbon chemicals. It has the ability to accumulate unsaturated 18 carbon chain length fatty acids, with additional minor contributions of saturated 18 carbon and 16 carbon fatty acids. | [8,9,10] |
| Cryptococcus curvatus | C. curvatus is oleaginous yeast capable of accumulating 18 carbon chain length fatty acids while growing on low or negative cost feedstock. Thus, it is a potential candidate for the use in industrial fermentation processes. In a rare case C. curvatus was found to be involved in peritonitis associated with gastric lymphoma. | [8,11,12] |
| Naganishia vishniacii (formerly known as Cryptococcus vishniacii) | N. vishniacii is psychrophilic yeast adapted to live in extreme conditions, such as low-temperature oligotrophic deserts. It also has the ability to grow in a low-nutrient environment, without added vitamins. | [8,13,14] |
| Cryptococcus wieringae | This is associated with pectin hydrolysis during the dew-wetting process of flax and found at the beginning of grape wine fermentation. | [8,15] |
| Cryptococcus amylolentus CBS 6273 | C. amylolentus is the most closely known related species of the pathogenic Cryptococcus species complex, and is non-pathogenic. | [7,16] |
| Kockovaella imperatae NRRL Y-17943 | K. imperatae is a non-pathogenic fungus used in the analysis of widespread adenine N6-methylation of active genes in fungal species. | [17] |
| Naematella encephela UCDFST | It is a parasite of another fungus, Stereum sanguinolentum. This fungus’ genome sequencing was carried out for the analysis of widespread adenine N6-methylation of active genes in fungal species. | [17] |
| Trichosporon asahii | Some species belonging to the genus Trichosporon are considered emerging opportunistic human pathogens and are the third most commonly isolated non-Candida yeasts from humans. They live in soil and are adapted to colonize the skin, gastrointestinal, respiratory and urinary tracts of humans. T. asahii is the most important species causing disseminated disease in immunocompromised patients, while the inhalation of T. asahii spores is the most important cause of summer-type hypersensitivity pneumonitis in healthy individuals. Some Trichosporon species have also emerged as rare but frequently fatal pathogens causing disseminated infections (trichosporonosis) in immunocompromised individuals and intensive care unit patients. | [18,19,20] |
| Trichosporon oleaginosus IBC0246 | T. oleaginosus is oleaginous yeast with the ability to accumulate lipids equivalent to biosynthetic kerosene, and thus is a biotechnologically valuable player for the generation of environmentally friendly (carbon-neutral) energy by converting agro-industrial waste to fuel (biodiesel). | [8,21] |
| Tremella mesenterica Fries | It is a parasite of crust fungus of the genus Peniophora and has a false appearance, as if it were growing on wood. Whereas in fact, it grows on the crust of fungal mycelium. | [22] |
| Species Name | Database | Reference |
|---|---|---|
| Cryptococcus gattii VGII R265 | NCBI | [3,4] |
| Cryptococcus gattii NT-10 | ||
| Cryptococcus gattii VGII 99/473 | ||
| Cryptococcus gattii E566 | ||
| Cryptococcus gattii VGII 2001/935-1 | ||
| Cryptococcus gattii VGIV IND107 | ||
| Cryptococcus gattii VGII CBS 10090 | ||
| Cryptococcus gattii VGII 2001/935-1 | ||
| Cryptococcus gattii EJB2 | ||
| Cryptococcus gattii WM276 | NCBI | [3] |
| Cryptococcus terricola JCM 24523 v1.0 | JGI | [9] |
| Cryptococcus curvatus ATCC 20509 | JGI | [11] |
| Naganishia vishniacii v1.0 (formerly known as Cryptococcus vishniacii) | JGI | |
| Cryptococcus wieringae | JGI | |
| Cryptococcus neoformans var. neoformans B-3501A | NCBI | [2] |
| Cryptococcus neoformans var. neoformans JEC21 | NCBI | [2] |
| Cryptococcus amylolentus CBS 6273 | JGI | [7,16] |
| Kockovaella imperatae NRRL Y-17943 v1.0 | JGI | [17] |
| Naematella encephela UCDFST 68-887.2 v1.0 | JGI | [17] |
| Trichosporon asahii var. asahii CBS 2479 | JGI | [19] |
| Trichosporon asahii var. asahii CBS 8904 | JGI | [20] |
| Trichosporon oleaginosus IBC0246 v1.0 | JGI | [21] |
| Tremella mesenterica Fries v1.0 | JGI | [22] |
| Cryptococcus neoformans var. grubii H99 | NCBI | [2] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akapo, O.O.; Padayachee, T.; Chen, W.; Kappo, A.P.; Yu, J.-H.; Nelson, D.R.; Syed, K. Distribution and Diversity of Cytochrome P450 Monooxygenases in the Fungal Class Tremellomycetes. Int. J. Mol. Sci. 2019, 20, 2889. https://doi.org/10.3390/ijms20122889
Akapo OO, Padayachee T, Chen W, Kappo AP, Yu J-H, Nelson DR, Syed K. Distribution and Diversity of Cytochrome P450 Monooxygenases in the Fungal Class Tremellomycetes. International Journal of Molecular Sciences. 2019; 20(12):2889. https://doi.org/10.3390/ijms20122889
Chicago/Turabian StyleAkapo, Olufunmilayo Olukemi, Tiara Padayachee, Wanping Chen, Abidemi Paul Kappo, Jae-Hyuk Yu, David R. Nelson, and Khajamohiddin Syed. 2019. "Distribution and Diversity of Cytochrome P450 Monooxygenases in the Fungal Class Tremellomycetes" International Journal of Molecular Sciences 20, no. 12: 2889. https://doi.org/10.3390/ijms20122889
APA StyleAkapo, O. O., Padayachee, T., Chen, W., Kappo, A. P., Yu, J. -H., Nelson, D. R., & Syed, K. (2019). Distribution and Diversity of Cytochrome P450 Monooxygenases in the Fungal Class Tremellomycetes. International Journal of Molecular Sciences, 20(12), 2889. https://doi.org/10.3390/ijms20122889