Nuclear-Biased DUSP6 Expression is Associated with Cancer Spreading Including Brain Metastasis in Triple-Negative Breast Cancer
Abstract
:1. Introduction
2. Results
2.1. HER2-Positive and HER2-Negative CTCs Detected and Isolated from Primary TNBC Patients
2.2. Distinct ERK1/2-MAPK Pathway Gene Profiles in HER2-Positive CTCs and HER2-Negative CTCs
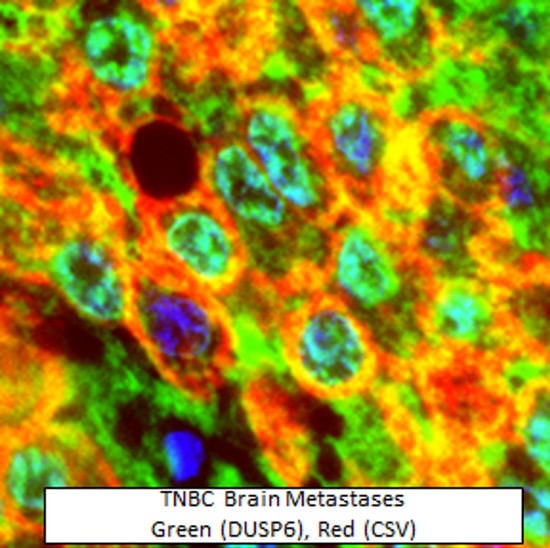
2.3. Nuclear Distribution of DUSP6 in CTCs and Brain Metastases from TNBC Patients
2.4. Nuclear Distribution of DUSP6 in Primary Tumor Sections in the Chemotherapy or Immunotherapy Treated 4T1 Metastatic Mouse Model
2.5. Novel Inhibition of Nuclear DUSP6 Mediated the Expression of ABCB5 and CSV In Vitro
2.6. Nuclear Association of DUSP6 with P300 and H3K9me2.
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Tissues
4.3. Animal Studies
4.4. Ethics
4.5. CTC Enrichment
4.6. CTC Detection, Imaging, and Isolation
4.7. RNA Isolation, cDNA Synthesis and Pre-Amplification from CTCs
4.8. Nanostring Analysis
4.9. ASI Microscopy
4.10. Nuclear to Cytoplasmic Fluorescence Ratio (Fn/c) Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three letter acronym |
| LD | linear dichroism |
| LSD1 | Lysine-specific histone demethylase 1 |
References
- Lin, N.U.; Vanderplas, A.; Hughes, M.E.; Theriault, R.L.; Edge, S.B.; Wong, Y.N.; Blayney, D.W.; Niland, J.C.; Winer, E.P.; Weeks, J.C. Clinicopathologic features, patterns of recurrence, and survival among women with triple-negative breast cancer in the national comprehensive cancer network. Cancer 2012, 118, 5463–5472. [Google Scholar] [CrossRef] [PubMed]
- Prasanna, T.; Wu, F.; Khanna, K.K.; Yip, D.; Malik, L.; Dahlstrom, J.E.; Rao, S. Optimizing poly (adp-ribose) polymerase inhibition through combined epigenetic and immunotherapy. Cancer Sci. 2018, 109, 3383–3392. [Google Scholar] [CrossRef] [PubMed]
- Zafar, A.; Wu, F.; Hardy, K.; Li, J.; Tu, W.J.; McCuaig, R.; Harris, J.; Khanna, K.K.; Attema, J.; Gregory, P.A.; et al. Chromatinized protein kinase c-theta directly regulates inducible genes in epithelial to mesenchymal transition and breast cancer stem cells. Mol. Cell Biol. 2014, 34, 2961–2980. [Google Scholar] [CrossRef] [PubMed]
- Boulding, T.; Wu, F.; McCuaig, R.; Dunn, J.; Sutton, C.R.; Hardy, K.; Tu, W.; Bullman, A.; Yip, D.; Dahlstrom, J.E.; et al. Differential roles for dusp family members in epithelial-to-mesenchymal transition and cancer stem cell regulation in breast cancer. PLoS ONE 2016, 11, e0148065. [Google Scholar] [CrossRef] [PubMed]
- Boulding, T.; McCuaig, R.D.; Tan, A.; Hardy, K.; Wu, F.; Dunn, J.; Kalimutho, M.; Sutton, C.R.; Forwood, J.K.; Bert, A.G.; et al. Lsd1 activation promotes inducible emt programs and modulates the tumour microenvironment in breast cancer. Sci. Rep. 2018, 8, 73. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Gradilone, A.; Raimondi, C.; Nicolazzo, C.; Petracca, A.; Gandini, O.; Vincenzi, B.; Naso, G.; Agliano, A.M.; Cortesi, E.; Gazzaniga, P. Circulating tumour cells lacking cytokeratin in breast cancer: The importance of being mesenchymal. J. Cell Mol. Med. 2011, 15, 1066–1070. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Bischoff, F.; Ann Mayer, J.; Wong, K.; Pham, T.; Kuerer, H.; Lodhi, A.; Bhattacharyya, A.; Hall, C.; Lucci, A. Discordance in her2 gene amplification in circulating and disseminated tumor cells in patients with operable breast cancer. Cancer Med. 2013, 2, 226–233. [Google Scholar] [CrossRef]
- Azim, H.A., Jr.; Rothe, F.; Aura, C.M.; Bavington, M.; Maetens, M.; Rouas, G.; Gebhart, G.; Gamez, C.; Eidtmann, H.; Baselga, J.; et al. Circulating tumor cells and response to neoadjuvant paclitaxel and her2-targeted therapy: A sub-study from the neoaltto phase iii trial. Breast 2013, 22, 1060–1065. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef]
- Jordan, N.V.; Bardia, A.; Wittner, B.S.; Benes, C.; Ligorio, M.; Zheng, Y.; Yu, M.; Sundaresan, T.K.; Licausi, J.A.; Desai, R.; et al. Her2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 2016, 537, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, B.A.S.; Neugebauer, J.; Andergassen, U.; Melcher, C.; Schochter, F.; Mouarrawy, D.; Ziemendorff, G.; Clemens, M.; Abel, E.V.; Heinrich, G.; et al. The her2 phenotype of circulating tumor cells in her2-positive early breast cancer: A translational research project of a prospective randomized phase iii trial. PLoS ONE 2017, 12, e0173593. [Google Scholar] [CrossRef] [PubMed]
- Levitan, D. Breast cancer ctcs can flip between her2 negativity and positivity. Cancer Netw. 2016. Available online: https://www.cancernetwork.com/her2-positive-breast-cancer/breast-cancer-ctcs-can-flip-between-her2-negativity-and-positivity (accessed on 20 September 2016).
- Buiga, P.; Elson, A.; Tabernero, L.; Schwartz, J.M. Regulation of dual specificity phosphatases in breast cancer during initial treatment with herceptin: A boolean model analysis. BMC Syst. Biol. 2018, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Cejudo-Marin, R.; Tarrega, C.; Nunes-Xavier, C.E.; Pulido, R. Caspase-3 cleavage of dusp6/mkp3 at the interdomain region generates active mkp3 fragments that regulate erk1/2 subcellular localization and function. J. Mol. Biol. 2012, 420, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Mathers, J.; Dickinson, R.J.; Mandl, M.; Keyse, S.M. Both nuclear-cytoplasmic shuttling of the dual specificity phosphatase mkp-3 and its ability to anchor map kinase in the cytoplasm are mediated by a conserved nuclear export signal. J. Biol Chem. 2004, 279, 41882–41891. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.K.; Abdollah, N.A.; Shafie, N.H.; Yusof, N.M.; Razak, S.R.A. Dual-specificity phosphatase 6 (dusp6): A review of its molecular characteristics and clinical relevance in cancer. Cancer Biol. Med. 2018, 15, 14–28. [Google Scholar] [PubMed]
- Arkell, R.S.; Dickinson, R.J.; Squires, M.; Hayat, S.; Keyse, S.M.; Cook, S.J. Dusp6/mkp-3 inactivates erk1/2 but fails to bind and inactivate erk5. Cell Signal. 2008, 20, 836–843. [Google Scholar] [CrossRef]
- Lonne, G.K.; Masoumi, K.C.; Lennartsson, J.; Larsson, C. Protein kinase cdelta supports survival of mda-mb-231 breast cancer cells by suppressing the erk1/2 pathway. J. Biol. Chem. 2009, 284, 33456–33465. [Google Scholar] [CrossRef]
- Nunes-Xavier, C.E.; Tarrega, C.; Cejudo-Marin, R.; Frijhoff, J.; Sandin, A.; Ostman, A.; Pulido, R. Differential up-regulation of map kinase phosphatases mkp3/dusp6 and dusp5 by ets2 and c-jun converge in the control of the growth arrest versus proliferation response of mcf-7 breast cancer cells to phorbol ester. J. Biol. Chem. 2010, 285, 26417–26430. [Google Scholar] [CrossRef]
- Song, H.; Wu, C.; Wei, C.; Li, D.; Hua, K.; Song, J.; Xu, H.; Chen, L.; Fang, L. Silencing of dusp6 gene by rnai-mediation inhibits proliferation and growth in mda-mb-231 breast cancer cells: An in vitro study. Int J. Clin. Exp. Med. 2015, 8, 10481–10490. [Google Scholar] [PubMed]
- Hagan, C.R.; Knutson, T.P.; Lange, C.A. A common docking domain in progesterone receptor-b links dusp6 and ck2 signaling to proliferative transcriptional programs in breast cancer cells. Nucleic Acids Res. 2013, 41, 8926–8942. [Google Scholar] [CrossRef] [PubMed]
- Lucci, M.A.; Orlandi, R.; Triulzi, T.; Tagliabue, E.; Balsari, A.; Villa-Moruzzi, E. Expression profile of tyrosine phosphatases in her2 breast cancer cells and tumors. Cell Oncol. 2010, 32, 361–372. [Google Scholar] [PubMed]
- Cui, Y.; Parra, I.; Zhang, M.; Hilsenbeck, S.G.; Tsimelzon, A.; Furukawa, T.; Horii, A.; Zhang, Z.Y.; Nicholson, R.I.; Fuqua, S.A. Elevated expression of mitogen-activated protein kinase phosphatase 3 in breast tumors: A mechanism of tamoxifen resistance. Cancer Res. 2006, 66, 5950–5959. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, F.; McCuaig, R.D.; Sutton, C.R.; Tan, A.H.Y.; Jeelall, Y.; Bean, E.G.; Dai, J.; Prasanna, T.; Batham, J.; Malik, L.; et al. Nuclear-Biased DUSP6 Expression is Associated with Cancer Spreading Including Brain Metastasis in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2019, 20, 3080. https://doi.org/10.3390/ijms20123080
Wu F, McCuaig RD, Sutton CR, Tan AHY, Jeelall Y, Bean EG, Dai J, Prasanna T, Batham J, Malik L, et al. Nuclear-Biased DUSP6 Expression is Associated with Cancer Spreading Including Brain Metastasis in Triple-Negative Breast Cancer. International Journal of Molecular Sciences. 2019; 20(12):3080. https://doi.org/10.3390/ijms20123080
Chicago/Turabian StyleWu, Fan, Robert D. McCuaig, Christopher R. Sutton, Abel H. Y. Tan, Yoshni Jeelall, Elaine G. Bean, Jin Dai, Thiru Prasanna, Jacob Batham, Laeeq Malik, and et al. 2019. "Nuclear-Biased DUSP6 Expression is Associated with Cancer Spreading Including Brain Metastasis in Triple-Negative Breast Cancer" International Journal of Molecular Sciences 20, no. 12: 3080. https://doi.org/10.3390/ijms20123080
APA StyleWu, F., McCuaig, R. D., Sutton, C. R., Tan, A. H. Y., Jeelall, Y., Bean, E. G., Dai, J., Prasanna, T., Batham, J., Malik, L., Yip, D., Dahlstrom, J. E., & Rao, S. (2019). Nuclear-Biased DUSP6 Expression is Associated with Cancer Spreading Including Brain Metastasis in Triple-Negative Breast Cancer. International Journal of Molecular Sciences, 20(12), 3080. https://doi.org/10.3390/ijms20123080