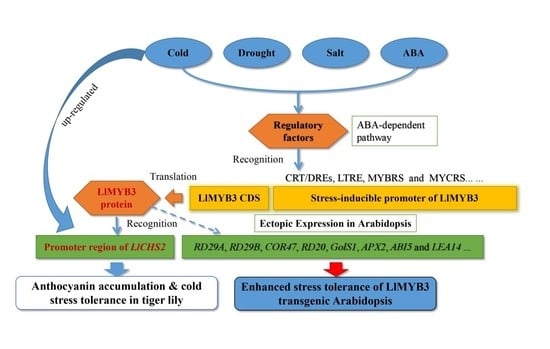
A MYB-Related Transcription Factor from Lilium lancifolium L. (LlMYB3) Is Involved in Anthocyanin Biosynthesis Pathway and Enhances Multiple Abiotic Stress Tolerance in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
2.1. Isolation of LlMYB3 and Sequence Analysis
2.2. Subcellular Localization and Transactivation Assay of LlMYB3
2.3. Expression Patterns of LlMYB3 under Multiple Stresses and ABA
2.4. Promoter Analysis of LlMYB3 under Multiple Stresses and ABA
2.5. Overexpression of LlMYB3 in Arabidopsis Improves Tolerance to Cold and Drought Stresses
2.6. Overexpression of LlMYB3 in Arabidopsis Increases Seed Sensitivity to ABA and Tolerance to NaCl
2.7. Altered Expression of Stress-Responsive Genes in LlMYB3 Transgenic Plants
2.8. LlMYB3 Can Bind to the Promoter of LlCHS2
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Cloning and Sequence Analysis of LlMYB3
4.3. Abiotic Stresses Treatment and Quantitative Real-Time PCR Analysis
4.4. Subcellular Localization and Transactivation Assay
4.5. Yeast One-Hybrid (Y1H) Assays
4.6. Generation of Transgenic Arabidopsis
4.7. Histochemical Staining and Fluorometric GUS Assay
4.8. Evaluation of Transgenic Plants Abiotic Stress Tolerance and ABA Sensitivity
4.9. Measurements of Relative Electrolyte Leakage, Soluble Sugar, and Water Loss Rate
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABA | abscisic acid |
| HCL | hierarchical clustering HCL |
| GFP | Green fluorescent protein |
| AbA | Aureobasidin A |
| GUS | β-glucuronidase |
| CHS | chalcone synthase |
| CHI | chalcone isomerase |
| F3H | flavanone 3-hydroxylase |
| F3’H | flavonoid 3′-hydroxylase |
| FLS | flavonol synthase |
| DFR | late biosynthetic genes dihydroflavonol reductase |
| LDOX/ANS | leucoanthocyanidin dioxygenase/anthocyanidin synthase |
| PAL1 | phenylalanine ammonia-lyase 1 |
| ANR2 | anthocyanidin reductase2 |
References
- Huang, G.T.; Ma, S.L.; Bai, L.P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ng, C.K.Y.; Fan, L.M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Yanhui, C.; Xiaoyuan, Y.; Kun, H.; Meihua, L.; Jigang, L.; Zhaofeng, G.; Zhiqiang, L.; Yunfei, Z.; Xiaoxiao, W.; Xiaoming, Q.; et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, H.; Ali, J.; Li, Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 2014, 9, e92913. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wang, Y.B.; Xie, Y.; Liang, Z.; Jiang, S.J.; Zhang, S.S.; Huang, Y.B.; Tang, Y.X. Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 2013, 20, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhangliang, C.; Kang, J.; Kang, D.; Gu, H.; Qin, G. AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol. Biol. Rep. 2013, 31, 87–97. [Google Scholar] [CrossRef]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.-H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2007, 281, 37636–37645. [Google Scholar] [CrossRef]
- Cui, M.H.; Yoo, K.S.; Hyoung, S.; Nguyen, H.T.K.; Kim, Y.Y.; Kim, H.J.; Ok, S.H.; Yoo, S.D.; Shin, J.S. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013, 587, 1773–1778. [Google Scholar] [CrossRef]
- Jung, C.; Seo, J.S.; Won Han, S.; Koo, Y.; Ho Kim, C.; Ik Song, S.; Hie Nahm, B.; Do Choi, Y.; Cheong, J.J. Overexpression of AtMYB44 Enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef]
- Abe, H. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Na Lee, Y.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Li, S.; An, X.; Liu, X.; Huanju, Q.; Wang, D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genom. 2009, 36, 17–29. [Google Scholar] [CrossRef]
- Pasquali, G.; Biricolti, S.; Locatelli, F.; Baldoni, E.; Mattana, M. OsMYB4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep. 2008, 27, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Vannini, C.; Campa, M.; Iriti, M.; Genga, A.; Faoro, F.; Carravieri, S.; Rotino, G.; Rossoni, M.; Spinardi, A.; Bracale, M. Evaluation of transgenic tomato plants ectopically expressing the rice Osmyb4 gene. Plant Sci. 2007, 173, 231–239. [Google Scholar] [CrossRef]
- Vannini, C.; Locatelli, F.; Bracale, M.; Magnani, E.; Marsoni, M.; Osnato, M.; Mattana, M.; Baldoni, E.; Coraggio, I. Overexpression of the rice OsMYB4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 2004, 37, 115–127. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Mao, X.; Jia, D.; Li, A.; Zhang, H.; Tian, S.; Zhang, X.; Jia, J.; Jing, R. Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis. Funct. Integr. Genom. 2011, 11, 445. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, M.; Tian, Y.; He, W.; Han, L.; Xia, G. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 2012, 39, 7183–7192. [Google Scholar] [CrossRef]
- Li, M.J.; Qiao, Y.; Li, Y.Q.; Shi, Z.L.; Zhang, N.; Bi, C.; Guo, J.K. A R2R3-MYB transcription factor gene in common wheat (namely TaMYBsm1) involved in enhancement of drought tolerance in transgenic Arabidopsis. J. Plant Res. 2016, 129. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, G.; Xia, C.; Jia, J.; Liu, X.; Kong, X. A wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. J. Exp. Bot. 2012, 63, 5873–5885. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, W.; Lv, J.; Jia, Y.; Wang, M.; Xia, G. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 2011, 63, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, G.; Zhao, G.; Xia, C.; Jia, J.; Liu, X.; Kong, X. Characterization of a wheat R2R3-MYB transcription factor gene, TaMYB19, involved in enhanced abiotic stresses in Arabidopsis. Plant Cell Physiol. 2014, 55. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 2019, 136, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Leyva, A.; Jarillo, J.A.; Salinas, J.; Martinez-Zapater, J.M. Low Temperature induces the accumulation of phenylalanine ammonia-lyase and chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiol. 1995, 108, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Rowan, D.; Cao, M.; Lin-Wang, K.; Cooney, J.; Jensen, D.; Austin, P.; B Hunt, M.; Norling, C.; Hellens, R.; J Schaffer, R.; et al. Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 2009, 182, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by over accumulation of antioxidant flavonoids. Plant J. 2013, 77. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Micheletti, D.; Palmer, J.; Volz, R.; Lozano, L.; Espley, R.; Hellens, R.; Chagne, D.; Rowan, D.; Troggio, M.; et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef]
- Li, L.; Ban, Z.; Li, X.H.; Wu, M.Y.; Wang, A.L.; Jiang, Y.; Jiang, Y.H. Differential expression of anthocyanin biosynthetic genes and transcription factor PcMYB10 in pears (Pyrus communis L.). PLoS ONE 2012, 7, e46070. [Google Scholar] [CrossRef]
- Ahmed, N.U.; Park, J.I.; Jung, H.J.; Hur, Y.; Nou, I.S. Anthocyanin biosynthesis for cold and freezing stress tolerance and desirable color in Brassica rapa. Funct. Integr. Genom. 2015, 15, 383–394. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Q.; Yang, Y.; Liu, X.; Gu, J.; Li, W.; Ma, S.; Lu, Y. De novo assembly and characterization of stress transcriptome and regulatory networks under temperature, salt and hormone stresses in Lilium lancifolium. Mol. Biol. Rep. 2014, 41, 8231–8245. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.B.; Li, W.Q.; Wang, J.M.; Zhang, Y.; Lu, Y.M. Identification of gene co-expression networks involved in cold resistance of Lilium lancifolium. Biol. Plant 2018, 62, 287–298. [Google Scholar] [CrossRef]
- Huang, P.; Chen, H.; Mu, R.; Yuan, X.; Zhang, H.S.; Huang, J. OsMYB511 encodes a MYB domain transcription activator early regulated by abiotic stress in rice. Genet. Mol. Res. 2015, 14, 9506–9517. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Huang, R.; Liu, Z.; Zhang, G. Functional characterization of cis-elements conferring vascular vein expression of At4g34880 amidase family protein gene in Arabidopsis. PLoS ONE 2013, 8, e67562. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Bai, X.; Zhu, Y.; Li, Y.; Cai, H.; Ji, W.; Ji, Z.; Liu, X.; Liu, X.; Li, J. A single-repeat R3-MYB transcription factor MYBC1 negatively regulates freezing tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 2010, 394, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- Su, C.F.; Wang, Y.C.; Hsieh, T.H.; Lu, C.A.; Tseng, T.H.; Yu, S.M. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 2010, 153, 145–158. [Google Scholar] [CrossRef]
- Shin, D.; Moon, S.J.; Han, S.; Kim, B.G.; Park, S.R.; Lee, S.K.; Yoon, H.J.; Lee, H.E.; Kwon, H.B.; Baek, D.; et al. Expression of StMYB1R-1, a novel potato single MYB-like domain transcription factor, increases drought tolerance. Plant Physiol. 2011, 155, 421–432. [Google Scholar] [CrossRef]
- Ganesan, G.; Sankararamasubramanian, H.M.; Harikrishnan, M.; Parida, A.; Ashwin, G. A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. J. Exp. Bot. 2012, 63, 4549–4561. [Google Scholar] [CrossRef]
- Fang, Q.; Wang, Q.; Mao, H.; Xu, J.; Wang, Y.; Hu, H.; He, S.; Tu, J.; Cheng, C.; Tian, G.; et al. AtDIV2, an R-R-type MYB transcription factor of Arabidopsis, negatively regulates salt stress by modulating ABA signaling. Plant Cell Rep. 2018, 37, 1499–1511. [Google Scholar] [CrossRef]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.; Lamb, C. Activation Tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2001, 12, 2383–2394. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; M Leavitt, J.; Lloyd, A. Regulation of the anthocyanin biosynthetic pathway by the TTG1/BHLH/MYB transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Favory, J.J.; Gruber, H.; Bartelniewoehner, L.; Bartels, S.; Binkert, M.; Funk, M.; Weisshaar, B.; Ulm, R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2009, 33, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Kimura, S.; Demura, T.; Takeda, J.; Ozeki, Y. DcMYB1 acts as a transcriptional activator of the carrot phenylalanine ammonia-lyase Gene (DcPAL1) in response to elicitor treatment, UV-B irradiation and the dilution effect. Plant Mol. Biol. 2005, 59, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Mellway, R.D.; T Tran, L.; Prouse, M.B.; Campbell, M.M.; Peter Constabel, C. The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol. 2009, 150, 924–941. [Google Scholar] [CrossRef] [PubMed]
- Ithal, N.; Reddy, A.R. Rice flavonoid pathway genes, OsDfr and OsAns, are induced by dehydration, high salt and ABA, and contain stress responsive promoter elements that interact with the transcription activator, OsC1-MYB. Plant Sci. 2004, 166, 1505–1513. [Google Scholar] [CrossRef]
- Altschul, S.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Aligment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hall, B.G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef] [Green Version]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A Free, Open-Source System for Microarray Data Management and Analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bent, A. Arabidopsis thaliana floral dip transformation method. Methods Mol. Biol. 2006, 343, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.H.; Liu, J.; He, X.J.; Mu, R.-L.; Zhou, H.L.; Chen, S.Y.; Zhang, J.S. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007, 143, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, L.; Xia, C.; Zhao, G.; Jia, J.; Kong, X. The novel wheat transcription factor TaNAC47 enhances multiple abiotic stress tolerances in transgenic plants. Front. Plant Sci. 2015, 6, 1174. [Google Scholar] [CrossRef] [PubMed]







| Site Name | (Strand) Position | Sequence | Function |
|---|---|---|---|
| ARE | (+)1277 | TGGTTT | cis-acting regulatory element essential for the anaerobic induction |
| CRT/DRE | (+)1323; (−)44 | GTCGAC | Core CRT/DRE motif |
| LTRE | (+)54 | ACCGACA | Putative low temperature responsive element |
| Box I | (+)696 | TTTCAAA | light responsive element |
| MNF1 | (−)32 | GTGCCCTATA | light responsive element |
| MBRS | (+)85,039; (−)91 | CAACGG (T/A)AACCA | MYB binding site involved in drought-inducibility |
| MYC | (+)8,438,501,089; (−)5,251,051 | CAA(T/C/A)TG CAT(T/G)TG | MYC recognition site involved in cold and drought-inducibility |
| CGTCA-motif | (+)855; (−)1009 | CGTCA | cis-acting regulatory element involved in the MeJA-responsiveness |
| TGACG-motif | (+)360; (−)484 | TGACG | cis-acting regulatory element involved in the MeJA-responsiveness |
| ERE | (+)695 | ATTTCAAA | ethylene-responsive element |
| TATC-BOX | (+)1274 | TATCCCA | cis-acting element involved in gibberellin-responsiveness |
| TGA-element | (−)490 | AACGAC | auxin-responsive element |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yong, Y.; Zhang, Y.; Lyu, Y. A MYB-Related Transcription Factor from Lilium lancifolium L. (LlMYB3) Is Involved in Anthocyanin Biosynthesis Pathway and Enhances Multiple Abiotic Stress Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 3195. https://doi.org/10.3390/ijms20133195
Yong Y, Zhang Y, Lyu Y. A MYB-Related Transcription Factor from Lilium lancifolium L. (LlMYB3) Is Involved in Anthocyanin Biosynthesis Pathway and Enhances Multiple Abiotic Stress Tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences. 2019; 20(13):3195. https://doi.org/10.3390/ijms20133195
Chicago/Turabian StyleYong, Yubing, Yue Zhang, and Yingmin Lyu. 2019. "A MYB-Related Transcription Factor from Lilium lancifolium L. (LlMYB3) Is Involved in Anthocyanin Biosynthesis Pathway and Enhances Multiple Abiotic Stress Tolerance in Arabidopsis thaliana" International Journal of Molecular Sciences 20, no. 13: 3195. https://doi.org/10.3390/ijms20133195
APA StyleYong, Y., Zhang, Y., & Lyu, Y. (2019). A MYB-Related Transcription Factor from Lilium lancifolium L. (LlMYB3) Is Involved in Anthocyanin Biosynthesis Pathway and Enhances Multiple Abiotic Stress Tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences, 20(13), 3195. https://doi.org/10.3390/ijms20133195