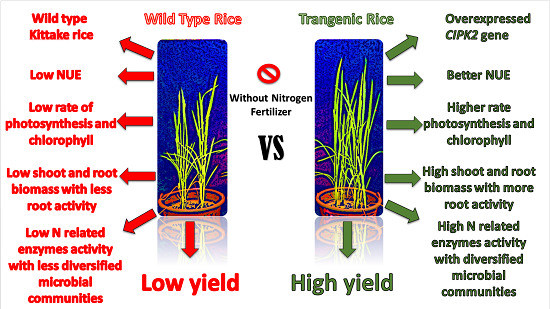
Exploring the Potential of Overexpressed OsCIPK2 Rice as a Nitrogen Utilization Efficient Crop and Analysis of Its Associated Rhizo-Compartmental Microbial Communities
Abstract
:1. Introduction
2. Results
2.1. Determination of CIPK2 Gene Expresion in Roots and Leaves of Transgenic and Wild-Type Rice
Expression of CIPK2 Protein in Roots and Leaves of RC and WT Rice
2.2. Bucket Experiment to Determine Differences between WT and RC
2.2.1. Changes in Tiller and Panicle
2.2.2. Chlorophyll (SPAD Values) and Changes in Photosynthetic Rate
2.2.3. Changes in Root Morphology
2.2.4. Root Activity
2.2.5. Changes in Nitrogen-Related Enzyme Activities in WT and RC
2.2.6. Changes in Dry Matter Weight and Root:Shoot Ratio
2.2.7. Changes in Yield and Yield Components
2.2.8. Changes in Nitrogen Use Efficiency
2.3. Changes in Bacterial Communities in Rhizosphere, Rhizoplane, and Endosphere of Rice Expressing CIPK2 with Enhanced Root-Specific Expression
2.3.1. Analysis of Bacterial Community Diversity
2.3.2. PCA Analysis of Rhizosphere Bacterial Community in Rice
2.3.3. Analysis of Bacterial Community Composition
2.3.4. Bacterial Functional Analysis
2.3.5. qPCR Verified the Changes of Genes Related to Rhizosphere Nitrogen Cycle under LN
2.4. Changes in Soil N-Related Physicochemical Properties under LN
2.5. The Influx and Efflux of Ca2+, NO3−, and NH4+ in WT and RC
3. Discussion
4. Methods
4.1. Plant Material and Transgenic Line Generation
4.2. Verification of CIPK2 Gene Overexpression in Rice Roots Thorough qPCR and Western Blot
4.2.1. qPCR
4.2.2. Western Blot
4.3. Rice Bucket Planting with Different N Treatments
4.3.1. Determination of Rice Plant Nitrogen and Nitrogen Utilization Ratio
4.3.2. Physiological Parameters
4.3.3. Determination of Rice-Related Enzymes in the Rice
4.4. Diversity of Bacterial Communities in Rhizosphere, Rhizoplane, and Endosphere of Rice
4.4.1. Sample Collection
4.4.2. Microbial DNA Extraction
4.4.3. Bacterium 16S rRNA Gene V3–V4 Region Amplification, Quantification, and Sequencing
4.4.4. Statistical and Sequencing Data Analysis
4.5. Quantitative Analysis of N Cycle Related Genes
4.6. Soil Physicochemical Properties and Enzyme Assays
4.7. Determination of Ion Flux Rate, Sample Preparation, and Determination Method
Data processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Amaral, L.R.; Molin, J.P.; Schepers, J.S. Algorithm for variable-rate nitrogen application in sugarcane based on active crop canopy sensor. Agron. J. 2015, 107, 1513–1523. [Google Scholar] [CrossRef]
- Khush, G.S. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 2005, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Good, A.G.; Beatty, P.H. Fertilizing nature: A tragedy of excess in the commons. PLoS Biol. 2011, 9, e1001124. [Google Scholar] [CrossRef] [PubMed]
- FAO. World Fertilizer Trends and Outlook to 2018. Food and Agriculture Organization of the United Nations (FAO): Rome, 2015. Available online: http://www.fao.org/3/a-i4324e.pdf.
- Hakeem, K.R.; Ahmad, A.; Iqbal, M.; Gucel, S.; Ozturk, M. Nitrogen-efficient rice cultivars can reduce nitrate pollution. Environ. Sci. Pollut. Res. 2011, 18, 1184–1193. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Buresh, R.J.; Huang, J.; Zhong, X.; Zou, Y.; Yang, J.; Wang, G.; Liu, Y.; Hu, R.; Tang, Q. Improving nitrogen fertilization in rice by sitespecific n management. A review. Agron. Sustain. Dev. 2010, 30, 649–656. [Google Scholar] [CrossRef]
- Huang, S.; Zhao, C.; Zhang, Y.; Wang, C. Nitrogen Use Efficiency in Rice. In Nitrogen in Agriculture-Updates; IntechOpen: London, UK, 2017; pp. 188–208. [Google Scholar]
- Pathak, R.R.; Ahmad, A.; Lochab, S.; Raghuram, N. Molecular physiology of plant nitrogen use efficiency and biotechnological options for its enhancement. Curr. Sci. 2008, 94, 1394–1403. [Google Scholar]
- Waqas, M.; Faheem, M.; Khan, A.; Shehzad, M.; Ansari, M. Estimation of heritability and genetic advance for some yield traits in eight f2 populations of wheat (Triticum aestivum l). Sci. Lett. 2014, 2, 43–47. [Google Scholar]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. The regulatory region controlling the nitrate-responsive expression of a nitrate reductase gene, nia1, in arabidopsis. Plant Cell Physiol. 2011, 52, 824–836. [Google Scholar] [CrossRef] [PubMed]
- McAllister, C.H.; Beatty, P.H.; Good, A.G. Engineering nitrogen use efficient crop plants: The current status. Plant Biotechnol. J. 2012, 10, 1011–1025. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.J.; Fan, X.; Orsel, M.; Smith, S.J.; Wells, D.M. Nitrate transport and signalling. J. Exp. Bot. 2007, 58, 2297–2306. [Google Scholar] [CrossRef] [PubMed]
- Suojalehto, H.; Kinaret, P.; Kilpeläinen, M.; Toskala, E.; Ahonen, N.; Wolff, H.; Alenius, H.; Puustinen, A. Level of fatty acid binding protein 5 (fabp5) is increased in sputum of allergic asthmatics and links to airway remodeling and inflammation. PLoS ONE 2015, 10, e0127003. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hu, B.; Yuan, D.; Liu, Y.; Che, R.; Hu, Y.; Ou, S.; Liu, Y.; Zhang, Z.; Wang, H. Expression of the nitrate transporter gene osnrt1. 1a/osnpf6. 3 confers high yield and early maturation in rice. Plant Cell 2018, 30, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Tang, Z.; Tan, Y.; Zhang, Y.; Luo, B.; Yang, M.; Lian, X.; Shen, Q.; Miller, A.J.; Xu, G. Overexpression of a ph-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. 2016, 113, 7118–7123. [Google Scholar] [CrossRef] [PubMed]
- Kudla, J.; Xu, Q.; Harter, K.; Gruissem, W.; Luan, S. Genes for calcineurin b-like proteins in arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. 1999, 96, 4718–4723. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.; Pelloux, J.; Brownlee, C.; Harper, J.F. Calcium at the crossroads of signaling. Plant Cell 2002, 14, S401–S417. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, J.; Wei, J.; Wang, H.; Wang, Y.; Ma, R. Functions and mechanisms of the cbl–cipk signaling system in plant response to abiotic stress. Prog. Nat. Sci. 2009, 19, 667–676. [Google Scholar] [CrossRef]
- Pandey, G.K.; Cheong, Y.H.; Kim, B.-G.; Grant, J.J.; Li, L.; Luan, S. Cipk9: A calcium sensor-interacting protein kinase required for low-potassium tolerance in arabidopsis. Cell Res. 2007, 17, 411. [Google Scholar] [CrossRef]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of stress-responsive cipk genes in rice for stress tolerance improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef]
- Wu, L.; Chen, J.; Wu, H.; Qin, X.; Wang, J.; Wu, Y.; Khan, M.U.; Lin, S.; Xiao, Z.; Luo, X. Insights into the regulation of rhizosphere bacterial communities by application of bio-organic fertilizer in pseudostellaria heterophylla monoculture regime. Front. Microbiol. 2016, 7, 1788. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Santos-Medellín, C.; Edwards, J.; Liechty, Z.; Nguyen, B.; Sundaresan, V. Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. MBio 2017, 8, e00764-17. [Google Scholar] [CrossRef] [PubMed]
- Schlaeppi, K.; Dombrowski, N.; Oter, R.G.; van Themaat, E.V.L.; Schulze-Lefert, P. Quantitative divergence of the bacterial root microbiota in arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. 2014, 111, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.B.; Vogel, C.; Bai, Y.; Vorholt, J.A. The plant microbiota: Systems-level insights and perspectives. Annu. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Bin, C.; Si-Ping, Z.; Li-Juan, Z.; Zhi-Min, L.; Ya-Na, S.; Wei-Wen, Z. Genetic diversity analysis of diazotrophs in the rice rhizosphere. Chin. J. Agric. Biotechnol. 2007, 4, 253–258. [Google Scholar] [CrossRef]
- Duan, Y.; Zhang, Y.; Ye, L.; Fan, X.; Xu, G.; Shen, Q. Responses of rice cultivars with different nitrogen use efficiency to partial nitrate nutrition. Ann. Bot. 2007, 99, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Moldenhauer, K.; Slaton, N. Rice growth and development. Rice Prod. Handb. 2001, 192, 7–14. [Google Scholar]
- Hai, B.; Diallo, N.H.; Sall, S.; Haesler, F.; Schauss, K.; Bonzi, M.; Assigbetse, K.; Chotte, J.-L.; Munch, J.C.; Schloter, M. Quantification of key genes steering the microbial nitrogen cycle in the rhizosphere of sorghum cultivars in tropical agroecosystems. Appl. Environ. Microbiol. 2009, 75, 4993–5000. [Google Scholar] [CrossRef]
- Song, W.; Makeen, K.; Wang, D.; Zhang, C.; Xu, Y.; Zhao, H.; Tu, E.; Zhang, Y.; Shen, Q.; Xu, G. Nitrate supply affects root growth differentially in two rice cultivars differing in nitrogen use efficiency. Plant Soil 2011, 343, 357–368. [Google Scholar] [CrossRef]
- Kan, C.-C.; Chung, T.-Y.; Juo, Y.-A.; Hsieh, M.-H. Glutamine rapidly induces the expression of key transcription factor genes involved in nitrogen and stress responses in rice roots. BMC Genom. 2015, 16, 731. [Google Scholar] [CrossRef] [PubMed]
- Miranda, R.d.S.; Alvarez-Pizarro, J.C.; Costa, J.H.; Paula, S.d.O.; Prisco, J.T.; Gomes-Filho, E. Putative role of glutamine in the activation of cbl/cipk signalling pathways during salt stress in sorghum. Plant Signal. Behav. 2017, 12, 522–536. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ju, X.; Zhang, F.; Pan, J.; Christie, P. Nitrogen dynamics and budgets in a winter wheat–maize cropping system in the north china plain. Field Crop. Res. 2003, 83, 111–124. [Google Scholar] [CrossRef]
- Waqas, M.; Feng, S.; Amjad, H.; Letuma, P.; Zhan, W.; Li, Z.; Fang, C.; Arafat, Y.; Khan, M.; Tayyab, M. Protein phosphatase (pp2c9) induces protein expression differentially to mediate nitrogen utilization efficiency in rice under nitrogen-deficient condition. Int. J. Mol. Sci. 2018, 19, 2827. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Y.; Hsu, P.-K.; Tsay, Y.-F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Ahkami, A.H.; White III, R.A.; Handakumbura, P.P.; Jansson, C. Rhizosphere engineering: Enhancing sustainable plant ecosystem productivity. Rhizosphere 2017, 3, 233–243. [Google Scholar] [CrossRef]
- Wu, L.; Chen, J.; Xiao, Z.; Zhu, X.; Wang, J.; Wu, H.; Wu, Y.; Zhang, Z.; Lin, W. Barcoded pyrosequencing reveals a shift in the bacterial community in the rhizosphere and rhizoplane of rehmannia glutinosa under consecutive monoculture. Int. J. Mol. Sci. 2018, 19, 850. [Google Scholar] [CrossRef] [PubMed]
- Knauth, S.; Hurek, T.; Brar, D.; Reinhold-Hurek, B. Influence of different oryza cultivars on expression of nifh gene pools in roots of rice. Environ. Microbiol. 2005, 7, 1725–1733. [Google Scholar] [CrossRef]
- Song, T.; Mårtensson, L.; Eriksson, T.; Zheng, W.; Rasmussen, U. Biodiversity and seasonal variation of the cyanobacterial assemblage in a rice paddy field in fujian, china. FEMS Microbiol. Ecol. 2005, 54, 131–140. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, Q.; Li, Z.; Cheng, W.; Sun, J.; Guo, Z.; Li, Y.; Zhou, J.; Meng, D.; Li, H. Environmental factors shaping the diversity of bacterial communities that promote rice production. BMC Microbiol. 2018, 18, 51. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichorst, S.A.; Trojan, D.; Roux, S.; Herbold, C.; Rattei, T.; Woebken, D. Genomic insights into the acidobacteria reveal strategies for their success in terrestrial environments. Environ. Microbiol. 2018, 20, 1041–1063. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.B.; Brady, S.M. Plant developmental responses to climate change. Dev. Biol. 2016, 419, 64–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiaoming, J.; Wenying, M.; Zeshu, Q. Species and enumeration of nitrogen-fixing bacteria in rice root systems. Acta Agric. Univ. Zhejiangensis 1989, 1, 57–60. [Google Scholar]
- Zehr, J.P. Nitrogen fixation by marine cyanobacteria. Trends Microbiol. 2011, 19, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Bothe, H.; Jost, G.; Schloter, M.; Ward, B.B.; Witzel, K.-P. Molecular analysis of ammonia oxidation and denitrification in natural environments. FEMS Microbiol. Rev. 2000, 24, 673–690. [Google Scholar] [CrossRef]
- Liu, W.; Lu, H.H.; Wu, W.; Wei, Q.K.; Chen, Y.X.; Thies, J.E. Transgenic bt rice does not affect enzyme activities and microbial composition in the rhizosphere during crop development. Soil Biol. Biochem. 2008, 40, 475–486. [Google Scholar] [CrossRef]
- Sun, H.; Li, J.; Song, W.; Tao, J.; Huang, S.; Chen, S.; Hou, M.; Xu, G.; Zhang, Y. Nitric oxide generated by nitrate reductase increases nitrogen uptake capacity by inducing lateral root formation and inorganic nitrogen uptake under partial nitrate nutrition in rice. J. Exp. Bot. 2015, 66, 2449–2459. [Google Scholar] [CrossRef] [Green Version]
- Lynch, J. Root architecture and plant productivity. Plant Physiol. 1995, 109, 7. [Google Scholar] [CrossRef]
- Ruffel, S.; Krouk, G.; Ristova, D.; Shasha, D.; Birnbaum, K.D.; Coruzzi, G.M. Nitrogen economics of root foraging: Transitive closure of the nitrate–cytokinin relay and distinct systemic signaling for n supply vs. Demand. Proc. Natl. Acad. Sci. 2011, 108, 18524–18529. [Google Scholar] [CrossRef]
- Sorgonà, A.; Lupini, A.; Abenavoli, M. Nitrate use-efficiency: A morphological analysis of the above-and below-ground functional traits in two citrus rootstocks. Glob. J. Plant Ecophysiol. 2011, 1, 26–37. [Google Scholar]
- Chen, H.; Li, D.; Zhao, J.; Xiao, K.; Wang, K. Effects of nitrogen addition on activities of soil nitrogen acquisition enzymes: A meta-analysis. Agric. Ecosyst. Environ. 2018, 252, 126–131. [Google Scholar] [CrossRef]
- Hu, H.C.; Wang, Y.Y.; Tsay, Y.F. Atcipk8, a cbl-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 2009, 57, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H.; Kobayashi, K.; Deji, A.; Sugiyama, T. Partial characterization of the signaling pathway for the nitrate-dependent expression of genes for nitrogen-assimilatory enzymes using detached maize leaves. Plant Cell Physiol. 1997, 38, 837–843. [Google Scholar] [CrossRef]
- Wang, R.; Okamoto, M.; Xing, X.; Crawford, N.M. Microarray analysis of the nitrate response in arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003, 132, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Kronzucker, H.J.; Siddiqi, M.Y.; Glass, A.D.; Kirk, G.J. Nitrate-ammonium synergism in rice. A subcellular flux analysis. Plant Physiol. 1999, 119, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 2008, 3, 824. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Xiong, D.; Chen, J.; Yu, T.; Gao, W.; Ling, X.; Li, Y.; Peng, S.; Huang, J. Spad-based leaf nitrogen estimation is impacted by environmental factors and crop leaf characteristics. Sci. Rep. 2015, 5, 13389. [Google Scholar] [CrossRef]
- Riaz, M.; Wu, X.; Yan, L.; Hussain, S.; Aziz, O.; Shah, A.; Jiang, C. Boron supply alleviates al-induced inhibition of root elongation and physiological characteristics in rapeseed (brassica napus l.). J. Plant Interact. 2018, 13, 270–276. [Google Scholar] [CrossRef]
- Gu, D.; Tang, L.; Cao, W.; Zhu, Y. Quantitative analysis on root morphological characteristics based on image analysis method in rice. Acta Agron. Sin. 2010, 36, 810–817. [Google Scholar] [CrossRef]
- Yu, X.-Z.; Zhang, F.-Z. Activities of nitrate reductase and glutamine synthetase in rice seedlings during cyanide metabolism. J. Hazard. Mater. 2012, 225, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than blast. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M. Soil Chemical Analysis; Prentice hall Inc.: Englewood Cliffs, NJ, USA, 1958; p. 498. [Google Scholar]
- Watanabe, F.; Olsen, S. Test of an ascorbic acid method for determining phosphorus in water and nahco3 extracts from soil 1. Soil Sci. Soc. Am. J. 1965, 29, 677–678. [Google Scholar] [CrossRef]
- Wang, Q.-Y.; Zhou, D.-M.; Cang, L. Microbial and enzyme properties of apple orchard soil as affected by long-term application of copper fungicide. Soil Biol. Biochem. 2009, 41, 1504–1509. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, A.; Li, X.; Kou, M.; Wang, W.; Chen, X.; Xu, T.; Zhu, M.; Ma, D.; Li, Z. Melatonin-stimulated triacylglycerol breakdown and energy turnover under salinity stress contributes to the maintenance of plasma membrane h+–atpase activity and K+/Na+ homeostasis in sweet potato. Front. Plant Sci. 2018, 9, 256. [Google Scholar] [CrossRef]







| Treatment | Genotype | Valid Panicles (pot−1) | Spikelets Per Panicle | Seed Setting Rate (%) | 1000 Grain Weight (g) | Grain Yield (g/pot−1) |
|---|---|---|---|---|---|---|
| LN | WT | 10.68c | 37b | 97.01a | 25a | 10.16c |
| RC | 14.67b | 43.25a | 82.08c | 25.28a | 13.75b | |
| NN | WT | 44a | 38.95a | 87.5b | 24.72a | 37.61a |
| RC | 44.67a | 38.46a | 89.82b | 24.63a | 37.43a |
| Treatment | Genotype | N Absorption (mg/plant) | Dry Matter Production Efficiency (BNUE) | Grain Production Efficiency (GNUE) | N Harvest Index (NHI) | Physiological Efficiency (PE) | Agronomic Utilization Rate (AE) | Absorption Utilization Rate (RE) |
|---|---|---|---|---|---|---|---|---|
| LN | WT | 42.28c | 128.59a | 63.14a | 0.65a | - | - | - |
| RC | 58.56b | 128.31a | 62.01a | 0.62b | - | - | - | |
| NN | WT | 190.56a | 93.65b | 50.29b | 0.49c | 46.26a | 13.26a | 28.68a |
| RC | 195.21a | 94.02b | 50.37b | 0.48c | 44.42b | 11.74b | 26.43b |
| Treatment | Sample | Observed_Species | Chao1 | Shannon | PD_Whole_Tree |
|---|---|---|---|---|---|
| No nitrogen | WS-L | 1833.97b | 2258.28a | 9.3032ab | 151.27b |
| RS-L | 1906.87a | 2327.28a | 9.4471a | 159.03a | |
| WP-L | 1721.50c | 2140.65b | 9.0611b | 144.92c | |
| RP-L | 1842.43b | 2296.41a | 9.2915ab | 156.69a | |
| WE-L | 672.13d | 1085.61c | 3.6811c | 75.16d | |
| RE-L | 645.20d | 1034.97c | 3.5526c | 74.35d | |
| Normal nitrogen | WS-N | 2119.53a | 2829.45a | 9.47a | 182.14a |
| RS-N | 2168.83a | 2894.73a | 9.47a | 190.34a | |
| WP-N | 2112.13a | 2799.20a | 9.47a | 183.68a | |
| RP-N | 2006.36a | 2719.17a | 9.24a | 178.34a | |
| WE-N | 764.56b | 1362.97b | 3.56b | 91.1b | |
| RE-N | 681.55b | 1219.97b | 2.94b | 85.56b |
| Gene | nifH | amoA (AOA) | amoA (AOB) | narG | nirK | nosZ | nirS |
|---|---|---|---|---|---|---|---|
| WT | 7.29 × 1012b | 4.99 × 107b | 4.20 × 106b | 1.69 × 106a | 7.59 ×1 04a | 1.77 × 106a | 9.01 × 107b |
| RC | 8.54 × 1012a | 5.60 × 107a | 4.75 × 106a | 1.83 × 106a | 3.50 × 104b | 2.83 × 106a | 12.84 × 107a |
| Genotype | NH4+-N (mg·kg−1) | NO3−-N (mg·kg−1) | Urease (mg·g−1·24h−1) | Invertase (mg·g−1·24h−1) | Nitrate Reductase (μg NO2−·g−1·24h−1) |
|---|---|---|---|---|---|
| WT | 27.52 ± 0.46a | 2.14 ± 0.60a | 0.118 ± 0.003b | 2.40 ± 0.083b | 2.52 ± 0.63b |
| RC | 24.85 ± 1.10b | 2.35 ± 0.49a | 0.138 ± 0.004a | 2.61 ± 0.062a | 5.53 ± 0.82a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.U.; Li, P.; Amjad, H.; Khan, A.Q.; Arafat, Y.; Waqas, M.; Li, Z.; Noman, A.; Islam, W.; Wu, L.; et al. Exploring the Potential of Overexpressed OsCIPK2 Rice as a Nitrogen Utilization Efficient Crop and Analysis of Its Associated Rhizo-Compartmental Microbial Communities. Int. J. Mol. Sci. 2019, 20, 3636. https://doi.org/10.3390/ijms20153636
Khan MU, Li P, Amjad H, Khan AQ, Arafat Y, Waqas M, Li Z, Noman A, Islam W, Wu L, et al. Exploring the Potential of Overexpressed OsCIPK2 Rice as a Nitrogen Utilization Efficient Crop and Analysis of Its Associated Rhizo-Compartmental Microbial Communities. International Journal of Molecular Sciences. 2019; 20(15):3636. https://doi.org/10.3390/ijms20153636
Chicago/Turabian StyleKhan, Muhammad Umar, Penghui Li, Hira Amjad, Ali Qaiser Khan, Yasir Arafat, Muhammad Waqas, Zhong Li, Ali Noman, Waqar Islam, Linkun Wu, and et al. 2019. "Exploring the Potential of Overexpressed OsCIPK2 Rice as a Nitrogen Utilization Efficient Crop and Analysis of Its Associated Rhizo-Compartmental Microbial Communities" International Journal of Molecular Sciences 20, no. 15: 3636. https://doi.org/10.3390/ijms20153636
APA StyleKhan, M. U., Li, P., Amjad, H., Khan, A. Q., Arafat, Y., Waqas, M., Li, Z., Noman, A., Islam, W., Wu, L., Zhang, Z., & Lin, W. (2019). Exploring the Potential of Overexpressed OsCIPK2 Rice as a Nitrogen Utilization Efficient Crop and Analysis of Its Associated Rhizo-Compartmental Microbial Communities. International Journal of Molecular Sciences, 20(15), 3636. https://doi.org/10.3390/ijms20153636