Culture Dependent and Independent Analysis of Potential Probiotic Bacterial Genera and Species Present in the Phyllosphere of Raw Eaten Produce
Abstract
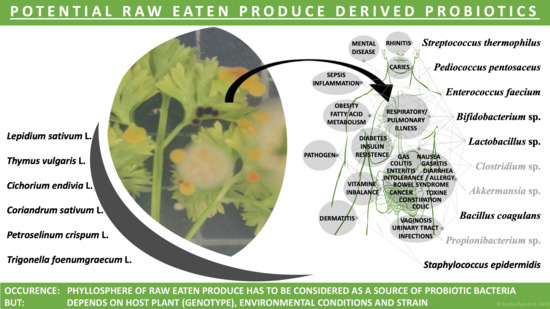
:1. Introduction
2. Results
2.1. Quantification of Potential Probiotic Bifidobacteria in the Phyllosphere of Edible Plants
2.2. Amount and Diversity of Cultivable Bacteria in Lepidium sativum, Cichorium endivia, and Thymus vulgaris
2.3. Isolation and Characterization of Phyllosphere Lactic Acid Bacteria (LAB)
2.4. Phyllosphere Microbiome Composition of Lepidium sativum
3. Discussion
4. Materials and Methods
4.1. Plant Species and Cultivation
4.2. Determination of Bifidobacterial-Specific Gene Copy Numbers Using Quantitative Real-Time PCR
4.3. Bacterial Cultivation
4.3.1. Total Cultivable Bacterial Numbers and Diversity on Complex Nutrient Agar
4.3.2. Specific Enrichment for Bifidobacteria and Lactobacilli
4.4. Protein Extraction from Bacterial Colonies for MALDI-TOF Biotyper Analysis
4.5. MALDI-TOF Mass Spectrometric Measurements and Isolate Identification
4.6. Plant Microbiome Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Liu, H.W.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.J.; Schenk, P.M. Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 2017, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.B.; Vogel, C.; Bai, Y.; Vorholt, J.A. The plant microbiota: Systems-level insights and perspectives. Annu. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Lebeis, S.L. Greater than the sum of their parts: Characterizing plant microbiomes at the community-level. Curr. Opin. Plant Biol. 2015, 24, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Andreote, F.D.; Silva, M. Microbial communities associated with plants: Learning from nature to apply it in agriculture. Curr. Opin. Microbiol. 2017, 37, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Hassani, M.A.; Duran, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Leach, J.E.; Triplett, L.R.; Argueso, C.T.; Trivedi, P. Communication in the phytobiome. Cell 2017, 169, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Remus-Emsermann, M.N.P.; Schlechter, R.O. Phyllosphere microbiology: At the interface between microbial individuals and the plant host. New Phytol. 2018, 218, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, G.; Coaker, G.L.; Leveau, J.H.J. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol. Lett. 2013, 348, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whipps, J.M.; Hand, P.; Pink, D.; Bending, G.D. Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 2008, 105, 1744–1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leff, J.W.; Fierer, N. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS ONE 2013, 8, e59310. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.R.; Randolph, K.C.; Osborn, S.L.; Tyler, H.L. Culture dependent and independent analysis of bacterial communities associated with commercial salad leaf vegetables. BMC Microbiol. 2013, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P.J.; Pink, D.A.C.; Bending, G.D. Cultivar-level genotype differences influence diversity and composition of lettuce (Lactuca sp.) phyllosphere fungal communities. Fungal Ecol. 2015, 17, 183–186. [Google Scholar] [CrossRef]
- Lopez-Velasco, G.; Carder, P.A.; Welbaum, G.E.; Ponder, M.A. Diversity of the spinach (Spinacia oleracea) spermosphere and phyllosphere bacterial communities. FEMS Microbiol. Lett. 2013, 346, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Gil, M.I.; Selma, M.V.; Suslow, T.; Jacxsens, L.; Uyttendaele, M.; Allende, A. Pre- and postharvest preventive measures and intervention strategies to control microbial food safety hazards of fresh leafy vegetables. Crit. Rev. Food Sci. Nutr. 2015, 55, 453–468. [Google Scholar] [CrossRef] [PubMed]
- Tatsika, S.; Karamanoli, K.; Karayanni, H.; Genitsaris, S. Metagenomic characterization of bacterial communities on ready-to-eat vegetables and effects of household washing on their diversity and composition. Pathogens 2019, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Ruppel, S. Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol. Ecol. 2014, 87, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Gorbach, S.L. Lactic acid bacteria and human health. Ann. Med. 1990, 22, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Hove, H.; Nørgaard, H.; Brøbech Mortensen, P. Lactic acid bacteria and the human gastrointestinal tract. Eur. J. Clin. Nutr. 1999, 53, 339–350. [Google Scholar] [CrossRef] [Green Version]
- Vaughan, E.E.; Heilig, H.G.H.J.; Ben-Amor, K.; de Vos, W.M. Diversity, vitality and activities of intestinal lactic acid bacteria and bifidobacteria assessed by molecular approaches. FEMS Microbiol. Rev. 2005, 29, 477–490. [Google Scholar] [CrossRef]
- Kerry, R.G.; Patra, J.K.; Gouda, S.; Park, Y.; Shin, H.-S.; Das, G. Benefaction of probiotics for human health: A review. J. Food Drug Anal. 2018, 26, 927–939. [Google Scholar] [CrossRef] [Green Version]
- McFarland, L.V.; Evans, C.T.; Goldstein, E.J.C. Strain-specificity and disease-specificity of probiotic efficacy: A systematic review and meta-analysis. Front. Med. 2018, 5, 124. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.C.; Kirjavainen, P.V.; Shortt, C.; Salminen, S. Probiotics: Mechanisms and established effects. Int. Dairy J. 1999, 9, 43–52. [Google Scholar] [CrossRef]
- Markowiak, P.; Slizewska, K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 2017, 9, 9. [Google Scholar]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Abriouel, H.; Lavilla Lerma, L.; Casado Munoz, M.C.; Perez Montoro, B.; Kabisch, J.; Pichner, R.; Cho, G.S.; Neve, H.; Fusco, V.; Franz, C.M.A.P.; et al. The controversial nature of the Weissella genus: Technological and functional aspects versus whole genome analysis-based pathogenic potential for their application in food and health. Front. Microbiol. 2015, 6, 1197. [Google Scholar] [CrossRef]
- Fusco, V.; Quero, G.M.; Cho, G.-S.; Kabisch, J.; Meske, D.; Neve, H.; Bockelmann, W.; Franz, C.M.A.P. The genus Weissella: Taxonomy, ecology and biotechnological potential. Front. Microbiol. 2015, 6, 155. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-J.; Lin, T.-L.; Tsai, Y.-L.; Wu, T.-R.; Lai, W.-F.; Lu, C.-C.; Lai, H.-C. Next generation probiotics in disease amelioration. J. Food Drug Anal. 2019, 3, 615–622. [Google Scholar] [CrossRef]
- Doron, S.; Snydman, D.R. Risk and safety of probiotics. Clin. Infect. Dis. 2015, 60, S129–S134. [Google Scholar] [CrossRef]
- Cassir, N.; Benamar, S.; La Scola, B. Clostridium butyricum: From beneficial to a new emerging pathogen. Clin. Microbiol. Infect. 2016, 22, 37–45. [Google Scholar] [CrossRef]
- Burton, J.P.; Wescombe, P.A.; Macklaim, J.M.; Chai, M.H.C.; MacDonald, K.; Hale, J.D.F.; Tagg, J.; Reid, G.; Gloor, G.B.; Cadieux, P.A. Persistence of the oral probiotic Streptococcus salivarius M18 is dose dependent and megaplasmid transfer can augment their bacteriocin production and adhesion characteristics. PLoS ONE 2013, 8, e65991. [Google Scholar] [CrossRef]
- Imperial, I.C.V.J.; Ibana, J.A. Addressing the antibiotic resistance problem with probiotics: Reducing the risk of its double-edged sword effect. Front. Microbiol. 2016, 7, 1983. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Zhang, R.; Tian, X.; Zhou, X.; Pan, X.; Wong, A. Assessing the risk of probiotic dietary supplements in the context of antibiotic resistance. Front. Microbiol. 2017, 8, 908. [Google Scholar] [CrossRef] [PubMed]
- Ayala, D.I.; Cook, P.W.; Franco, J.G.; Bugarel, M.; Kottapalli, K.R.; Loneragan, G.H.; Brashears, M.M.; Nightingale, K.K. A systematic approach to identify and characterize the effectiveness and safety of novel probiotic strains to control foodborne pathogens. Front. Microbiol. 2019, 10, 1108. [Google Scholar] [CrossRef] [PubMed]
- Pontonio, E.; Di Cagno, R.; Tarraf, W.; Filannino, P.; De Mastro, G.; Gobbetti, M. Dynamic and assembly of epiphyte and endophyte lactic acid bacteria during the life cycle of Origanum vulgare L. Front. Microbiol. 2018, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Savitha, Y.L.; Mulla, S.R.; Preethi, D.M.; Sandeep, C.; Suvarna, V.C.; Suresh, C.K. Molecular characterization of Lactobacillus plantarum isolates of vegetables and phyllosphere. J. Pure Appl. Microbiol. 2013, 7, 3201–3206. [Google Scholar]
- Zwielehner, J.; Handschur, M.; Michaelsen, A.; Irez, S.; Demel, M.; Denner, E.B.M.; Hasiberger, A.G. DGGE and real-time PCR analysis of lactic acid bacteria in bacterial communities of the phyllosphere of lettuce. Mol. Nutr. Food Res. 2008, 52, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Wassermann, B.; Rybakova, D.; Muller, C.; Berg, G. Harnessing the microbiomes of Brassica vegetables for health issues. Sci. Rep. 2017, 7, 12. [Google Scholar] [CrossRef]
- Martins, E.M.F.; Ramos, A.M.; Vanzela, E.S.L.; Stringheta, P.C.; de Oliveira Pinto, C.L.; Martins, J.M. Products of vegetable origin: A new alternative for the consumption of probiotic bacteria. Food Res. Int. 2013, 51, 764–770. [Google Scholar] [CrossRef]
- Montenegro, D.; Kalpana, K.; Chrissian, C.; Sharma, A.; Takaoka, A.; Iacovidou, M.; Soll, C.E.; Aminova, O.; Heguy, A.; Cohen, L.; et al. Uncovering potential ’herbal probiotics’ in Juzen-taiho-to through the study of associated bacterial populations. Bioorganic Med. Chem. Lett. 2015, 25, 466–469. [Google Scholar] [CrossRef]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Miyamoto, Y.; Takada, T.; Matsumoto, K.; Oyaizu, H.; Tanaka, R. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 2002, 68, 5445–5451. [Google Scholar] [CrossRef]
- Huschek, D.; Witzel, K. Rapid dereplication of microbial isolates using Matrix-Assisted Laser Desorption Ionization Time-Of-Flight Mass Spectrometry: A mini-review. J. Adv. Res. 2019, 19, 99–104. [Google Scholar] [CrossRef]
- Tapsell, L.C.; Hemphill, I.; Cobiac, L.; Sullivan, D.R.; Fenech, M.; Patch, C.S.; Roodenrys, S.; Keogh, J.; Clifton, P.; Williams, P. Health benefits of herbs and spices: The past, the present, the future. Med. J. Aust. 2006, 185, S1–S24. [Google Scholar]
- Moursi, H. Die Heilpflanzen im Land der Pharaonen; Eigenverlag: Kairo, Egypt, 1992; p. 372. [Google Scholar]
- Sakkas, H.; Papadopoulou, C. Antimicrobial activity of basil, oregano, and thyme essential oils. J. Microbiol. Biotechnol. 2017, 27, 429–438. [Google Scholar] [CrossRef] [PubMed]
- FAO/WHO. Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation. Available online: www.fao.org/3/a-a0512e.pdf (accessed on 16 July 2019).
- Gibson, G.R.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota—Introducing the concept of prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- De Vrese, M.; Schrezenmeir, J. Probiotics, Prebiotics, and Synbiotics. In Food Biotechnology; Stahl, U., Donalies, U.E.B., Nevoigt, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 1–66. [Google Scholar]
- Ruppel, S.; Rühlmann, J.; Merbach, W. Quantification and localization of bacteria in plant tissues using quantitative real-time PCR and online emission fingerprinting. Plant Soil 2006, 286, 21–35. [Google Scholar] [CrossRef]
- Akalın, A.S.; Fenderya, S.; Akbulut, N. Viability and activity of bifidobacteria in yoghurt containing fructooligosaccharide during refrigerated storage. Int. J. Food Sci. Technol. 2004, 39, 613–621. [Google Scholar] [CrossRef]
- Bull, M.; Plummer, S.; Marchesi, J.; Mahenthiralingam, E. The life history of Lactobacillus acidophilus as a probiotic: A tale of revisionary taxonomy, misidentification and commercial success. FEMS Microbiol. Lett. 2013, 349, 77–87. [Google Scholar] [CrossRef]
- Vemuri, R.; Shinde, T.; Shastri, M.D.; Perera, A.P.; Tristram, S.; Martoni, C.J.; Gundamaraju, R.; Ahuja, K.D.K.; Ball, M.; Eri, R. A human origin strain Lactobacillus acidophilus DDS-1 exhibits superior in vitro probiotic efficacy in comparison to plant or dairy origin probiotics. Int. J. Med Sci. 2018, 15, 840–848. [Google Scholar] [CrossRef]
- Chung, W.H.; Kang, J.; Lim, M.Y.; Lim, T.J.; Lim, S.; Roh, S.W.; Nam, Y.-D. Complete genome sequence and genomic characterization of Lactobacillus acidophilus LA1 (11869BP). Front. Pharmacol. 2018, 9, 7. [Google Scholar] [CrossRef]
- Furrie, E. Probiotics and allergy. Proc. Nutr. Soc. 2005, 64, 465–469. [Google Scholar] [CrossRef] [Green Version]
- Dixit, Y.; Wagle, A.; Vakil, B. Patents in the field of probiotics, prebiotics, synbiotics: A review. J. Food Microbiol. Saf. Hyg. 2016, 1, 1–13. [Google Scholar] [CrossRef]
- Foligne, B.; Daniel, C.; Pot, B. Probiotics from research to market: The possibilities, risks and challenges. Curr. Opin. Microbiol. 2013, 16, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Sarate, P.J.; Heinl, S.; Poiret, S.; Drinic, M.; Zwicker, C.; Schabussova, I.; Daniel, C.; Wiedermann, U. E. coli Nissle 1917 is a safe mucosal delivery vector for a birch-grass pollen chimera to prevent allergic poly-sensitization. Mucosal Immunol. 2019, 12, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Hoppe, M.; Onning, G.; Berggren, A.; Hulthen, L. Probiotic strain Lactobacillus plantarum 299v increases iron absorption from an iron-supplemented fruit drink: A double-isotope cross-over single-blind study in women of reproductive age. Br. J. Nutr. 2015, 114, 1195–1202. [Google Scholar] [CrossRef] [PubMed]
- Vasijevic, T.; Shah, N.P. Probiotics—From Metchnikoff to bioactives. Int. Dairy J. 2008, 18, 714–728. [Google Scholar] [CrossRef]
- Zielinska, D.; Kolozyn-Krajewska, D. Food-origin lactic acid bacteria may exhibit probiotic properties: Review. Biomed. Res. Int. 2018, 2018, 5063185. [Google Scholar] [CrossRef]
- Varankovich, N.V.; Nickerson, M.T.; Korber, D.R. Probiotic-based strategies for therapeutic and prophylactic use against multiple gastrointestinal diseases. Front. Microbiol. 2015, 6, 685. [Google Scholar] [CrossRef] [Green Version]
- Aureli, P.; Fiore, A.; Scalfaro, C.; Casale, M.; Franciosa, G. National survey outcomes on commercial probiotic food supplements in Italy. Int. J. Food Microbiol. 2010, 137, 265–273. [Google Scholar] [CrossRef]
- Tarrah, A.; de Castilhos, J.; Rossi, R.C.; Duarte, V.D.; Ziegler, D.R.; Corich, V.; Giacomini, A. In vitro probiotic potential and anti-cancer activity of newly isolated folate-producing Streptococcus thermophilus strains. Front. Microbiol. 2018, 9, 2214. [Google Scholar] [CrossRef]
- Han, S.H.; Suk, K.T.; Kim, D.J.; Kim, M.Y.; Baik, S.K.; Kim, Y.D.; Cheon, G.J.; Choi, D.H.; Ham, Y.L.; Shin, D.H.; et al. Effects of probiotics (cultured Lactobacillus subtilis/Streptococcus faecium) in the treatment of alcoholic hepatitis: Randomized-controlled multicenter study. Eur. J. Gastroenterol. Hepatol. 2015, 27, 1300–1306. [Google Scholar] [CrossRef]
- Teneva-Angelova, T.; Beshkova, D. Non-traditional sources for isolation of lactic acid bacteria. Ann. Microbiol. 2016, 66, 449–459. [Google Scholar] [CrossRef]
- Holzapfel, W.H.; Haberer, P.; Snel, J.; Schillinger, U.; Huis in’t Veld, J.H.J. Overview of gut flora and probiotics. Int. J. Food Microbiol. 1998, 41, 85–101. [Google Scholar] [CrossRef]
- Sharma, P.; Kaur, S.; Kaur, R.; Kaur, M.; Kaur, S. Proteinaceous secretory metabolites of probiotic human commensal Enterococcus hirae 20c, E. faecium 12a and L12b as antiproliferative agents against cancer cell lines. Front. Microbiol. 2018, 9, 948. [Google Scholar] [CrossRef] [PubMed]
- Mansour, N.M.; Heine, H.; Abdou, S.M.; Shenana, M.E.; Zakaria, M.K.; El-Diwany, A. Isolation of Enterococcus faecium NM113, Enterococcus faecium NM213 and Lactobacillus casei NM512 as novel probiotics with immunomodulatory properties. Microbiol. Immunol. 2014, 58, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Lathrop, A. Effects of Lactobacillus plantarum, Pediococcus acidilactici, and Pediococcus pentosaceus on the growth of Listeria monocytogenes and Salmonella on alfalfa sprouts. J. Food Prot. 2019, 82, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Lee, D.; Jang, I.S.; Choi, H.S.; Suh, H.J. The culture of Pediococcus pentosaceus T1 inhibits Listeria proliferation in salmon fillets and controls maturation of kimchi. Food Technol. Biotechnol. 2015, 53, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Jonganurakkun, B.; Wang, Q.; Xu, S.H.; Tada, Y.; Minamida, K.; Yasokawa, D.; Sugi, M.; Hara, H.; Asano, K. Pediococcus pentosaceus NB-17 for probiotic use. J. Biosci. Bioeng. 2008, 106, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Taroub, B.; Salma, L.; Manel, Z.; Ouzari, H.I.; Hamdi, Z.; Moktar, H. Isolation of lactic acid bacteria from grape fruit: Antifungal activities, probiotic properties, and in vitro detoxification of ochratoxin A. Ann. Microbiol. 2019, 69, 17–27. [Google Scholar] [CrossRef]
- Walther, B.; Schmid, A. Chapter 7—Effect of fermentation on vitamin content in food. In Fermented Foods in Health and Disease Prevention; Frias, J., Martinez-Villaluenga, C., Peñas, E., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 131–157. [Google Scholar]
- Duc, L.H.; Hong, H.A.; Barbosa, T.M.; Henriques, A.O.; Cutting, S.M. Characterization of Bacillus probiotics available for human use. Appl. Environ. Microbiol. 2004, 70, 2161–2171. [Google Scholar] [CrossRef]
- Elshaghabee, F.M.F.; Rokana, N.; Gulhane, R.D.; Sharma, C.; Panwar, H. Bacillus as potential probiotics: Status, concerns, and future perspectives. Front. Microbiol. 2017, 8, 1490. [Google Scholar] [CrossRef]
- El Hage, R.; Hernandez-Sanabria, E.; Van de Wiele, T. Emerging trends in “Smart Probiotics”: Functional consideration for the development of novel health and industrial applications. Front. Microbiol. 2017, 8, 1889. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Feng, S.; Arjan, N.; Chen, W. A next generation probiotic, Akkermansia muciniphila. Crit. Rev. Food Sci. Nutr. 2018, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Anhe, F.F.; Pilon, G.; Roy, D.; Desjardins, Y.; Levy, E.; Marette, A. Triggering Akkermansia with dietary polyphenols: A new weapon to combat the metabolic syndrome? Gut Microbes. 2016, 7, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.-J.; Marito, S.; Yang, J.-J.; Keshari, S.; Chew, C.-H.; Chen, C.-C.; Huang, C.-M. A Microtube Array Membrane (MTAM) encapsulated live fermenting Staphylococcus epidermidis as a skin probiotic patch against Cutibacterium acnes. Int. J. Mol. Sci. 2019, 20, 14. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.N.; Rea, M.C.; O’Connor, P.M.; Hill, C.; Ross, R.P. Human skin microbiota is a rich source of bacteriocin-producing Staphylococci that kill human pathogens. FEMS Microbiol. Ecol. 2019, 95, fiy241. [Google Scholar] [CrossRef] [PubMed]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef] [PubMed]
- Sung, C.; Kim, B.G.; Kim, S.; Joo, H.S.; Kim, P.I. Probiotic potential of Staphylococcus hominis MBBL 2–9 as anti-Staphylococcus aureus agent isolated from the vaginal microbiota of a healthy woman. J. Appl. Microbiol. 2010, 108, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Khusro, A.; Aarti, C.; Dusthackeer, A.; Agastian, P. Anti-tubercular and probiotic properties of coagulase-negative staphylococci isolated from Koozh, a traditional fermented food of South India. Microb. Pathog. 2018, 114, 239–250. [Google Scholar] [CrossRef]
- McGuire, M.K.; McGuire, M.A. Human milk: Mother nature’s prototypical probiotic food? Adv. Nutr. 2015, 6, 112–123. [Google Scholar] [CrossRef]
- Takahashi, M.; Taguchi, H.; Yamaguchi, H.; Osaki, T.; Komatsu, A.; Kamiya, S. The effect of probiotic treatment with Clostridium butyricum on enterohemorrhagic Escherichia coli O157:H7 infection in mice. FEMS Immunol. Med. Microbiol. 2004, 41, 219–226. [Google Scholar] [CrossRef]
- Holland, R.; Liu, S.Q. Lactic Acid Bacteria: Leuconostoc spp. In Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., Ed.; Academic Press: Cambridge, MA, USA, 2011; pp. 138–142. [Google Scholar]
- Müller, T.; Ulrich, A.; Ott, E.M.; Müller, M. Identification of plant-associated enterococci. J. Appl. Microbiol. 2001, 91, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Mundt, J.O.; Beattie, W.G.; Wieland, F.R. Pediococci residing on plants. J. Bacteriol. 1969, 98, 938–942. [Google Scholar]
- Konuray, G.; Erginkaya, Z. Potential use of Bacillus coagulans in the food industry. Foods 2018, 7, 92. [Google Scholar] [CrossRef] [PubMed]
- Sarhan, M.S.; Hamza, M.A.; Youssef, H.H.; Patz, S.; Becker, M.; ElSawey, H.; Nemr, R.; Daanaa, H.S.A.; Mourad, E.F.; Morsi, A.T.; et al. Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media—A review. J. Adv. Res. 2019, 19, 15–27. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis—The “accidental” pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [PubMed]




| Biodiversity Indices | L. sativum | C. endivia | T. vulgaris |
|---|---|---|---|
| N | 8018 | 56 | 276 |
| S | 28 | 26 | 30 |
| H | 1.95 | 2.87 | 2.72 |
| Hmax | 3.33 | 3.26 | 3.40 |
| E | 0.59 | 0.88 | 0.80 |
| PHYLA | ORDER | GENUS | SPECIES |
|---|---|---|---|
| Actinobacteria | Bifidobacteriales | Bifidobacteria | unclassified |
| Firmicutes | Lactobacillales | Lactobacillus | unclassified |
| Streptococcus | thermophilus | ||
| Streptococcus | unclassified |
| Probiotic Group | Species | Phyllosphere (Plant) | Probiotic Reference |
|---|---|---|---|
| Lactobacillus 1 | (Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae) | ||
| Lactobacillus sp. | LS | [50,51,52,53,54,55] | |
| L. plantarum | CS | [20,54,55,56,57,58,59] | |
| Bifidobacteria | (Bacteria; Terrabacteria group; Actinobacteria; Actinobacteria; Bifidobacteriales) | ||
| Bifidobacteria sp. | LS | [53,55,60] | |
| Streptococcus 1 | (Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae) | ||
| Streptococcus sp. | LS | [20,54,55] | |
| S. thermophilus | LS | [20,53,54,61,62] | |
| Enterococcus 1 | (Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Enterococcaceae) | ||
| E. faecium | CE, TF | [20,53,54,55,60,63,64,65,66,67] | |
| Pediococcus 1 | (Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae) | ||
| P. pentosaceus | CE, CS, PC, TV | [20,53,55,59,60,68,69,70,71] | |
| Leuconostoc1 | (Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Leuconostocaceae) | ||
| L. lactis | TF | [72] | |
| Bacillus | (Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae) | ||
| Bacillus spp. | LS, PC, TF, TV | [55,73,74] | |
| Propionibacterium | (Bacteria; Terrabacteria group; Actinobacteria; Actinobacteria; Propionibacteriales; Propionibacteriaceae) | ||
| Propionibacterium sp. | LS | [20,54,55] | |
| Akkermansia | (Bacteria; PVC group; Verrucomicrobia; Verrucomicrobiae; Verrucomicrobiales; Akkermansiaceae) | ||
| Akkermansia sp. | LS | [20,75,76,77] | |
| Staphylococcus | (Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Staphylococcaceae) | ||
| S. epidermidis | CS | [78,79,80] | |
| S. hominis | CS, TV | [80,81,82,83] | |
| Clostridium | (Bacteria; Terrabacteria group; Firmicutes; Clostridia; Clostridiales; Clostridiaceae) | ||
| Clostridium sp. | LS | [29,84] | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patz, S.; Witzel, K.; Scherwinski, A.-C.; Ruppel, S. Culture Dependent and Independent Analysis of Potential Probiotic Bacterial Genera and Species Present in the Phyllosphere of Raw Eaten Produce. Int. J. Mol. Sci. 2019, 20, 3661. https://doi.org/10.3390/ijms20153661
Patz S, Witzel K, Scherwinski A-C, Ruppel S. Culture Dependent and Independent Analysis of Potential Probiotic Bacterial Genera and Species Present in the Phyllosphere of Raw Eaten Produce. International Journal of Molecular Sciences. 2019; 20(15):3661. https://doi.org/10.3390/ijms20153661
Chicago/Turabian StylePatz, Sascha, Katja Witzel, Ann-Christin Scherwinski, and Silke Ruppel. 2019. "Culture Dependent and Independent Analysis of Potential Probiotic Bacterial Genera and Species Present in the Phyllosphere of Raw Eaten Produce" International Journal of Molecular Sciences 20, no. 15: 3661. https://doi.org/10.3390/ijms20153661
APA StylePatz, S., Witzel, K., Scherwinski, A. -C., & Ruppel, S. (2019). Culture Dependent and Independent Analysis of Potential Probiotic Bacterial Genera and Species Present in the Phyllosphere of Raw Eaten Produce. International Journal of Molecular Sciences, 20(15), 3661. https://doi.org/10.3390/ijms20153661