A Novel Effector Protein of Apple Proliferation Phytoplasma Disrupts Cell Integrity of Nicotiana spp. Protoplasts
Abstract
:1. Introduction
2. Results
2.1. In Silico Analysis of PME2 Indicates Effector Potential
2.2. Atp_00136 (Pme2) is Expressed in P. Mali-Infected Malus × Domestica but not in the Insect Vector C. Picta
2.3. Genetic Variability of Pme2
2.4. PME2ST and PME2AT Translocate to the Nucleus of Nicotiana spp. Protoplasts
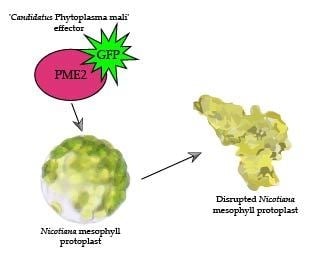
2.5. PME2ST but not PME2AT Affect Cell Integrity of Nicotiana spp. Protoplasts
2.6. A Yeast Two-Hybrid Screen Was Unsuitable for the Elucidation of PME2ST Function
3. Discussion
4. Materials and Methods
4.1. Verification of Pme2 Expression in Malus × Domestica and C. Picta
4.2. Amplification, Subcloning, and Sequencing of atp_00136
4.3. Subcloning of Pme2 into GreenGate Expression Vectors
4.4. Protoplast Isolation and Transformation
4.5. Nicotiana spp. Leaf Infiltration
4.6. Expression in Yeast
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AP | Apple proliferation |
| AY-WB | Aster Yellow Witches’ Broom |
| C. picta | Cacopsylla picta |
| FDA | Fluorescein diacetate |
| GFP | Green fluorescent protein |
| N. | Nicotiana |
| NES | Nuclear export signal |
| NLS | Nuclear localization signal |
| PI | Propidium iodide |
| P. mali | Candidatus Phytoplasma mali |
| PME2 | Protein in Malus Expressed 2 |
| qPCR | quantitative PCR |
| SNP | Single nucleotide polymorphism |
| Y2H | Yeast two-hybrid |
References
- Strauss, E. Phytoplasma research begins to bloom. Science 2009, 325, 388–390. [Google Scholar] [CrossRef]
- Christensen, N.M.; Axelsen, K.B.; Nicolaisen, M.; Schulz, A. Phytoplasmas and their interactions with hosts. Trends Plant Sci. 2005, 10, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Hogenhout, S.A.; Oshima, K.; Ammar, E.-D.; Kakizawa, S.; Kingdom, H.N.; Namba, S. Phytoplasmas: Bacteria that manipulate plants and insects. Mol. Plant Pathol. 2008, 9, 403–423. [Google Scholar] [CrossRef] [PubMed]
- Sugio, A.; MacLean, A.M.; Kingdom, H.N.; Grieve, V.M.; Manimekalai, R.; Hogenhout, S.A. Diverse targets of phytoplasma effectors: From plant development to defense against insects. Annu. Rev. Phytopathol. 2011, 49, 175–195. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Correa, V.R.; Toruño, T.Y.; Ammar, E.-D.; Kamoun, S.; Hogenhout, S.A. AY-WB phytoplasma secretes a protein that targets plant cell nuclei. Mol. Plant Microbe Interact. 2009, 22, 18–30. [Google Scholar] [CrossRef]
- Lu, Y.-T.; Cheng, K.-T.; Jiang, S.-Y.; Yang, J.-Y. Post-translational cleavage and self-interaction of the phytoplasma effector SAP11. Plant Signal. Behav. 2014, 9, e28991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.-T.; Li, M.-Y.; Cheng, K.-T.; Tan, C.M.; Su, L.-W.; Lin, W.-Y.; Shih, H.-T.; Chiou, T.-J.; Yang, J.-Y. Transgenic plants that express the phytoplasma effector SAP11 show altered phosphate starvation and defense responses. Plant Physiol. 2014, 164, 1456–1469. [Google Scholar] [CrossRef]
- Sugawara, K.; Honma, Y.; Komatsu, K.; Himeno, M.; Oshima, K.; Namba, S. The alteration of plant morphology by small peptides released from the proteolytic processing of the bacterial peptide TENGU. Plant Physiol. 2013, 162, 2005–2014. [Google Scholar] [CrossRef]
- MacLean, A.M.; Sugio, A.; Makarova, O.V.; Findlay, K.C.; Grieve, V.M.; Tóth, R.; Nicolaisen, M.; Hogenhout, S.A. Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol. 2011, 157, 831–841. [Google Scholar] [CrossRef]
- Sugio, A.; Kingdom, H.N.; MacLean, A.M.; Grieve, V.M.; Hogenhout, S.A. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, E1254–E1263. [Google Scholar] [CrossRef] [Green Version]
- Tomkins, M.; Kliot, A.; Marée, A.F.; Hogenhout, S.A. A multi-layered mechanistic modelling approach to understand how effector genes extend beyond phytoplasma to modulate plant hosts, insect vectors and the environment. Curr. Opin. Plant Biol. 2018, 44, 39–48. [Google Scholar] [CrossRef] [PubMed]
- MacLean, A.M.; Orlovskis, Z.; Kowitwanich, K.; Zdziarska, A.M.; Angenent, G.C.; Immink, R.G.H.; Hogenhout, S.A. Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol. 2014, 12, e1001835. [Google Scholar] [CrossRef] [PubMed]
- Janik, K.; Mithöfer, A.; Raffeiner, M.; Stellmach, H.; Hause, B.; Schlink, K.; Mithofer, A. An effector of apple proliferation phytoplasma targets TCP transcription factors-a generalized virulence strategy of phytoplasma? Mol. Plant Pathol. 2017, 18, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Strohmayer, A.; Moser, M.; Si-Ammour, A.; Krczal, G.; Boonrod, K. ’Candidatus Phytoplasma mali’ genome encodes a protein that functions as a E3 Ubiquitin Ligase and could inhibit plant basal defense. Mol. Plant Microbe Interact. 2019. [Google Scholar] [CrossRef] [PubMed]
- Boonrod, K.; Munteanu, B.; Jarausch, B.; Jarausch, W.; Krczal, G. An immunodominant membrane protein (Imp) of ’Candidatus Phytoplasma mali’ binds to plant actin. Mol. Plant Microbe Interact. 2012, 25, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Seemüller, E.; Kampmann, M.; Kiss, E.; Schneider, B. HflB gene-based phytopathogenic classification of ’Candidatus Phytoplasma mali’ strains and evidence that strain composition determines virulence in multiply infected apple trees. Mol. Plant Microbe Interact. 2011, 24, 1258–1266. [Google Scholar] [CrossRef]
- Schneider, B.; Sule, S.; Jelkmann, W.; Seemüller, E. Suppression of aggressive strains of ’Candidatus phytoplasma mali’ by mild strains in Catharanthus roseus and Nicotiana occidentalis and indication of similar action in apple trees. Phytopathology 2014, 104, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Seemüller, E.; Zikeli, K.; Furch, A.C.U.; Wensing, A.; Jelkmann, W. Virulence of ‘Candidatus Phytoplasma mali’ strains is closely linked to conserved substitutions in AAA+ ATPase AP460 and their supposed effect on enzyme function. Eur. J. Plant Pathol. 2017, 86, 141. [Google Scholar] [CrossRef]
- Kube, M.; Schneider, B.; Kuhl, H.; Dandekar, T.; Heitmann, K.; Migdoll, A.M.; Reinhardt, R.; Seemüller, E. The linear chromosome of the plant-pathogenic mycoplasma ’Candidatus Phytoplasma mali’. BMC Genomics 2008, 9, 306. [Google Scholar] [CrossRef]
- Kube, M.; Mitrovic, J.; Duduk, B.; Rabus, R.; Seemüller, E. Current view on phytoplasma genomes and encoded metabolism. Scientific World J. 2012, 2012, 185942. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Beckwith, J. The Sec-dependent pathway. Res. Microbiol. 2013, 164, 497–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Cour, T.; Kiemer, L.; Mølgaard, A.; Gupta, R.; Skriver, K.; Brunak, S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004, 17, 527–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-59259-890-8. [Google Scholar]
- Journal of Bacteriology. Nomenclature: Genetic Nomenclature. Available online: https://jb.asm.org/content/nomenclature (accessed on 16 September 2019).
- Luge, T.; Kube, M.; Freiwald, A.; Meierhofer, D.; Seemüller, E.; Sauer, S. Transcriptomics assisted proteomic analysis of Nicotiana occidentalis infected by Candidatus Phytoplasma mali strain AT. Proteomics 2014, 14, 1882–1889. [Google Scholar] [CrossRef] [PubMed]
- Hogenhout, S.A.; Van der Hoorn, R.A.L.; Terauchi, R.; Kamoun, S. Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 2009, 22, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, P.; Ahmed, B.; Joly, D.L.; Germain, H. Effector biology during biotrophic invasion of plant cells. Virulence 2014, 5, 703–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.; Liang, C.; Li, F.; Wang, L.; Wu, X.; Lu, A.; Xiao, G.; Zhang, G. The rules and functions of nucleocytoplasmic shuttling proteins. Int. J. Mol. Sci. 2018, 19, 1445. [Google Scholar] [CrossRef] [PubMed]
- Corbett, A.H.; Silver, P.A. Nucleocytoplasmic transport of macromolecules. Microbiol. Mol. Biol. Rev. 1997, 61, 193–211. [Google Scholar] [PubMed]
- Görlich, D.; Dabrowski, M.; Bischoff, F.R.; Kutay, U.; Bork, P.; Hartmann, E.; Prehn, S.; Izaurralde, E. A novel class of RanGTP binding proteins. J. Cell Biol 1997, 138, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Ward, B.; Medville, R.; Lazarowitz, S.-G.; Turgeon, R. The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J. Virol. 1997, 71, 3726–3733. [Google Scholar] [PubMed]
- Minato, N.; Himeno, M.; Hoshi, A.; Maejima, K.; Komatsu, K.; Takebayashi, Y.; Kasahara, H.; Yusa, A.; Yamaji, Y.; Oshima, K.; et al. The phytoplasmal virulence factor TENGU causes plant sterility by downregulating of the jasmonic acid and auxin pathways. Sci. Rep. 2014, 4, 7399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitazawa, Y.; Iwabuchi, N.; Himeno, M.; Sasano, M.; Koinuma, H.; Nijo, T.; Tomomitsu, T.; Yoshida, T.; Okano, Y.; Yoshikawa, N.; et al. Phytoplasma-conserved phyllogen proteins induce phyllody across the Plantae by degrading floral MADS domain proteins. J. Exp. Bot. 2017, 68, 2799–2811. [Google Scholar] [CrossRef] [PubMed]
- Noueiry, A.O.; Lucas, W.J.; Gilbertson, R.L. Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 1994, 76, 925–932. [Google Scholar] [CrossRef]
- Sanderfoot, A.A.; Ingham, D.J.; Lazarowitz, S.-G. A viral movement protein as a nuclear shuttle: The geminivirus BR1 movement protein contains domains essential for interaction with BL1 and nuclear localization. Plant Physiol. 1996, 110, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Sanderfoot, A.A.; Lazarowitz, S.-G. Cooperation in viral movement: The geminivirus BL1 movement protein interacts with BR1 and redirects it from the nucleus to the cell periphery. The Plant Cell 1995, 7, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Sanderfoot, A.A.; Lazarowitz, S.-G. Getting it together in plant virus movement: Cooperative interactions between bipartite geminivirus movement proteins. Trends Cell Biol. 1996, 6, 353–358. [Google Scholar] [CrossRef]
- Fields, S.; Song, O.-K. A novel genetic system to detect protein–protein interactions. Nature 1989, 340, 245–246. [Google Scholar] [CrossRef]
- Brückner, A.; Polge, C.; Lentze, N.; Auerbach, D.; Schlattner, U. Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci. 2009, 10, 2763–2788. [Google Scholar] [CrossRef] [PubMed]
- Musetti, R.; Paolacci, A.; Ciaffi, M.; Tanzarella, O.A.; Polizzotto, R.; Tubaro, F.; Mizzau, M.; Ermacora, P.; Badiani, M.; Osler, R. Phloem cytochemical modification and gene expression following the recovery of apple plants from apple proliferation disease. Phytopathology 2010, 100, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.R.; Schneider, B.; Mithöfer, A.; Reichelt, M.; Seemüller, E.; Furch, A.C.U. Implications of Candidatus Phytoplasma mali infection on phloem function of apple trees. Endocytobiosis Cell Res. 2015, 26, 67–75. [Google Scholar]
- Kay, S.; Hahn, S.; Marois, E.; Wieduwild, R.; Bonas, U. Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3Deltarep16. Plant J. 2009, 59, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Win, J.; Chaparro-Garcia, A.; Belhaj, K.; Saunders, D.G.O.; Yoshida, K.; Dong, S.; Schornack, S.; Zipfel, C.; Robatzek, S.; Hogenhout, S.A.; et al. Effector biology of plant-associated organisms: Concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.P.; Castañeda, A.; Zhao, G.; Erdos, G.; Gabriel, D.W. Expression of a single, host-specific, bacterial pathogenicity gene in plant cells elicits division, enlargement, and cell death. Mol. Plant Microbe Interact. 1999, 12, 556–560. [Google Scholar] [CrossRef]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef]
- Domingues, M.N.; de Souza, T.A.; Cernadas, R.A.; de Oliveira, M.L.P.; Docena, C.; Farah, C.S.; Benedetti, C.E. The Xanthomonas citri effector protein PthA interacts with citrus proteins involved in nuclear transport, protein folding and ubiquitination associated with DNA repair. Mol. Plant Pathol. 2010, 11, 663–675. [Google Scholar] [CrossRef]
- de Souza, T.A.; Soprano, A.S.; de Lira, N.P.V.; Quaresma, A.J.C.; Pauletti, B.-A.; Paes Leme, A.-F.; Benedetti, C.-E. The TAL effector PthA4 interacts with nuclear factors involved in RNA-dependent processes including a HMG protein that selectively binds poly(U) RNA. PLoS ONE 2012, 7, e32305. [Google Scholar] [CrossRef]
- Kay, S.; Hahn, S.; Marois, E.; Hause, G.; Bonas, U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 2007, 318, 648–651. [Google Scholar] [CrossRef]
- Seemüller, E.; Schneider, B. Differences in virulence and genomic features of strains of ’Candidatus Phytoplasma mali’, the apple proliferation agent. Phytopathology 2007, 97, 964–970. [Google Scholar] [CrossRef] [PubMed]
- Seemüller, E.; Kampmann, M.; Kiss, E.; Schneider, B. Molecular differentiation of severe and mild strains of ‘Candidatus Phytoplasma mali’ and evidence that their interaction in multiply infected trees determines disease severity. Bull. Insectology 2011, 64, 163–164. [Google Scholar]
- Rid, M.; Mesca, C.; Ayasse, M.; Gross, J. Apple proliferation phytoplasma influences the pattern of plant volatiles emitted depending on pathogen virulence. Front. Ecol. Evol. 2016, 3, 271. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Mittelberger, C.; Obkircher, L.; Oettl, S.; Oppedisano, T.; Pedrazzoli, F.; Panassiti, B.; Kerschbamer, C.; Anfora, G.; Janik, K. The insect vector Cacopsylla picta vertically transmits the bacterium ‘Candidatus Phytoplasma mali’ to its progeny. Plant Pathol 2017, 66, 1015–1021. [Google Scholar] [CrossRef]
- Monti, M.; Martini, M.; Tedeschi, R. EvaGreen Real-time PCR protocol for specific ’Candidatus Phytoplasma mali’ detection and quantification in insects. Mol. Cell. Probes 2013, 27, 129–136. [Google Scholar] [CrossRef]
- Lampropoulos, A.; Sutikovic, Z.; Wenzl, C.; Maegele, I.; Lohmann, J.U.; Forner, J. GreenGate - A novel, versatile, and efficient cloning system for plant transgenesis. PLoS ONE 2013, 8, e83043. [Google Scholar] [CrossRef]
- Janik, K.; Stellmach, H.; Mittelberger, C.; Hause, B. Characterization of phytoplasmal effector protein interaction with proteinaceous plant host targets using bimolecular fluorescence complementation (BiFC). In Phytoplasmas: Methods and Protocols; Musetti, R., Pagliari, L., Eds.; Humana Press: New York, NY, USA, 2019; pp. 321–331. ISBN 978-1-4939-8837-2. [Google Scholar]
- Heslop-Harrison, J.; Heslop-Harrison, Y. Evaluation of pollen viability by enzymatically induced fluorescence; intracellular hydrolysis of fluorescein diacetate. Stain Technol. 1970, 45, 115–120. [Google Scholar] [CrossRef]
- Janik, K.; Schlink, K. Unravelling the function of a bacterial effector from a non-cultivable plant pathogen using a yeast two-hybrid screen. J. Vis. Exp. 2017, 119, e55150. [Google Scholar] [CrossRef]







| Month | Status | Pool | cDNA Integrity (tip41) | Phytoplasma (16S) | atp_00136 |
|---|---|---|---|---|---|
| May | non-infected | 1 | 26.38 | N/A | N/A |
| 2 | 26.38 | N/A | N/A | ||
| Oct | non-infected | 3 | 26.58 | N/A | N/A |
| 2 | 26.59 | N/A | N/A | ||
| 3 | 26.58 | N/A | N/A | ||
| May | infected | 1 | 26.56 | N/A | N/A |
| 2 | 26.53 | N/A | N/A | ||
| 3 | 26.61 | N/A | N/A | ||
| Oct | infected | 1 | 26.71 | 23.67 | 28.00 |
| 2 | 26.44 | 23.18 | 27.66 | ||
| 3 | 26.48 | 23.34 | 28.34 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mittelberger, C.; Stellmach, H.; Hause, B.; Kerschbamer, C.; Schlink, K.; Letschka, T.; Janik, K. A Novel Effector Protein of Apple Proliferation Phytoplasma Disrupts Cell Integrity of Nicotiana spp. Protoplasts. Int. J. Mol. Sci. 2019, 20, 4613. https://doi.org/10.3390/ijms20184613
Mittelberger C, Stellmach H, Hause B, Kerschbamer C, Schlink K, Letschka T, Janik K. A Novel Effector Protein of Apple Proliferation Phytoplasma Disrupts Cell Integrity of Nicotiana spp. Protoplasts. International Journal of Molecular Sciences. 2019; 20(18):4613. https://doi.org/10.3390/ijms20184613
Chicago/Turabian StyleMittelberger, Cecilia, Hagen Stellmach, Bettina Hause, Christine Kerschbamer, Katja Schlink, Thomas Letschka, and Katrin Janik. 2019. "A Novel Effector Protein of Apple Proliferation Phytoplasma Disrupts Cell Integrity of Nicotiana spp. Protoplasts" International Journal of Molecular Sciences 20, no. 18: 4613. https://doi.org/10.3390/ijms20184613
APA StyleMittelberger, C., Stellmach, H., Hause, B., Kerschbamer, C., Schlink, K., Letschka, T., & Janik, K. (2019). A Novel Effector Protein of Apple Proliferation Phytoplasma Disrupts Cell Integrity of Nicotiana spp. Protoplasts. International Journal of Molecular Sciences, 20(18), 4613. https://doi.org/10.3390/ijms20184613