Non-Targeted Metabolomics Reveals Sorghum Rhizosphere-Associated Exudates are Influenced by the Belowground Interaction of Substrate and Sorghum Genotype
Abstract
:1. Introduction
2. Results
2.1. Soil Characteristics and Viable Microbial Presences Vary Among Substrates
2.2. Variation in Plant Morphology is Largely Influenced by Substrate
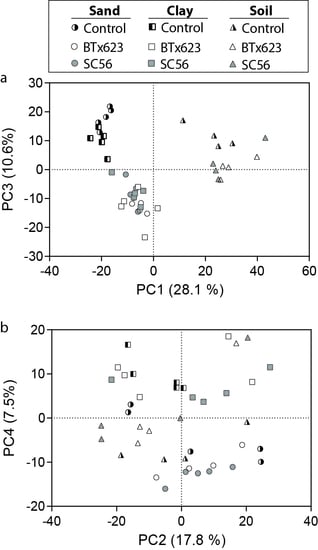
2.3. Non-Targeted Metabolomics Detected Rhizosphere-Enhanced or -Abated Metabolites
2.4. Annotated Metabolites Represent Known Root Exudates
3. Discussion
3.1. Rhizosphere-Associated Exudation Responds to Stressful Abiotic Conditions
3.2. Root Exudates Serve to Enlist Plant Growth-Promoting Bacteria
3.3. Metabolites Can Be Abated by the Rhizosphere Environment
3.4. Rhizosphere-Associated Metabolite Detection and Analysis Considerations
4. Materials and Methods
4.1. Plant Cultivation
4.2. Experimental Design
4.3. Characterization of Soil Properties and Quantitative Estimation of Viable Soil Microorganisms
4.4. Assessment of Morphological and Physiological Plant Traits
4.5. Metabolite Extraction
4.6. Metabolite Detection by Gas Chromatography—Mass Spectrometry
4.7. Metabolite Detection by Ultra Performance Liquid Chromatography—Mass Spectrometry
4.8. Metabolomics Data Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GC-MS | Gas Chromatography-Mass Spectrometry |
| HPLC | High Performance Liquid Chromatography |
| PGPM | Plant Growth-Promoting Microorganisms |
| REMs | Rhizosphere-Enhanced Metabolites |
| RAMs | Rhizosphere-Abated Metabolites |
| UPLC-MS | Ultra-Performance Liquid Chromatography-Mass Spectrometry |
References
- Bengough, A.G.; Bransby, M.F.; Hans, J.; McKenna, S.J.; Roberts, T.J. Root responses to soil physical conditions; growth dynamics from field to cell. J. Exp. Bot. 2006, 57, 437–447. [Google Scholar] [CrossRef]
- Bertin, C.; Yang, X.; Weston, L.A. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 2003, 256, 67–83. [Google Scholar] [CrossRef]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudate. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root exudation of phytochemicals in arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 2013, 8, e55731. [Google Scholar] [CrossRef]
- Jones, D.L.; Hodge, A.; Kuzyakov, Y. Plant and mycorrhizal regulation of rhizodeposition. New Phytol. 2004, 163, 459–480. [Google Scholar] [CrossRef] [Green Version]
- Bakker, M.G.; Manter, D.K.; Sheflin, A.M.; Weir, T.L.; Vivanco, J.M. Harnessing the rhizosphere microbiome through plant breeding and agricultural management. Plant Soil 2012, 360, 1–13. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fertil Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Kuijken, R.C.; van Eeuwijk, F.A.; Marcelis, L.F.; Bouwmeester, H.J. Root phenotyping: From component trait in the lab to breeding. J. Exp. Bot. 2015, 66, 5389–5401. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Frankenberger, W.T. Microbial biosynthesis of ethylene and its influence on plant growth. In Advances in Microbial Ecology; Marshall, K.C., Ed.; Springer: Boston, MA, USA, 1992; Volume 12. [Google Scholar]
- Kumar, V.V. Plant growth-promoting microorganisms. In Plant, Soil and Microbes; Hekeem, K.R., Akhtar, M.S., Abdullah, S.N.A., Eds.; Springer: New York, NY, USA, 2016; Volume 1, pp. 1–16. [Google Scholar]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Romheld, V. The Release of Root Exudates as Affected by the Plant Physiological Status. In The Rhizosphere: Biochemistry and Organic Substances at the Soil-Plant Interface; CRC Press: Boca Raton, FL, USA, 2007; Volume 2, pp. 23–72. [Google Scholar]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Neumann, G.; Bott, S.; Ohler, M.A.; MOck, H.P.; Lippmann, R.; Grosch, R.; Smalla, K. Root exudation and root development of lettuce (Lactuca sativa l. Cv. Tizian) as affected by different soils. Front. Microbiol. 2014, 5, 2. [Google Scholar] [CrossRef]
- Pierret, A.; Doussan, C.; Capowiez, Y.; Bastardie, F. Root functional architecture: A framework for modeling the interplay between roots and soil. Vadose Zone J. 2007, 6, 269–281. [Google Scholar] [CrossRef]
- Berns, A.E.; Philipp, H.; Narres, H.-D.; Burauel, P.; Vereecken, H.; Tappe, W. Effect of gamma-sterilization and autoclaving on soil organic matter structure as studied by solid state nmr, uv and fluorescence spectroscopy. Eur. J. Soil Sci. 2008, 59, 540–550. [Google Scholar] [CrossRef]
- Liegel, L.H. Effects of sterilization procedures on the biological, chemical, and physical properties of soils: A review. Turrialba 1986, 36, 11–19. [Google Scholar]
- Chairidchai, P.; Ritchie, G.S.P. Zinc adsorption by sterilized and non-sterilized soil in the presence of citrate and catechol. Commun. Soil Sci. Plant Anal. 1993, 24, 261–275. [Google Scholar] [CrossRef]
- Vranova, V.; Rejsek, K.; Skene, K.R.; Janous, D.; Formanek, P. Methods of collection of plant root exudates in relation to plant metabolism and purpose: A review. J. Plant Nutr. Soil Sci. 2013, 176, 175–199. [Google Scholar] [CrossRef]
- Dakora, F.D.; Phillips, D.A. Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 2002, 245, 35–47. [Google Scholar] [CrossRef]
- Curl, E.A.; Truelove, B. The Rhizosphere; Springer: Berlin, Germany, 1986; pp. 55–92. [Google Scholar]
- Czarnota, M.A.; Paul, R.N.; Dayan, R.E.; Nimbal, C.I.; Weston, L.A. Mode of action, localization of production, chemical nature, and activity of sorgoleone: A potent psii inhibitor in Sorghum spp. Root exudates. Weed Technol. 2003, 15, 813–825. [Google Scholar] [CrossRef]
- Dayan, F.E.; Kagan, I.A.; Rimando, A.M. Elucidation of the biosynthetic pathway of the allelochemical sorgoleone using retrobiosynthetic nmr analysis. J. Biol. Chem. 2003, 278, 28607–28611. [Google Scholar] [CrossRef] [PubMed]
- Netzley, D.H.; Butler, L.G. Roots of sorghum exude hydrophobic droplets containing biologically active components. Crop Sci. 1986, 26, 775–780. [Google Scholar] [CrossRef]
- Walker, T.S.; Bais, H.P.; Grotewold, E.; Vivanco, J.M. Root exudation and rhizosphere biology. Plant Physiol. 2003, 132, 44–51. [Google Scholar] [CrossRef]
- Heuberger, A.L.; Broeckling, C.D.; Kirkpatrick, K.R.; Prenni, J.E. Application of nontargeted metabolite profiling to discover novel markers of quality traits in an advanced population of malting barley. Plant Biotechnol. 2014, 12, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.F.; Heuberger, A.L.; Kirkwood, J.S.; Collins, C.C.; Wolfrum, E.J.; Broeckling, C.D.; Prenni, J.E.; Jahn, C.E. Non-targeted metabolomics in diverse sorghum breeding lines indicates primary and secondary metabolite profiles are associated with plant biomass accumuation and photosynthesis. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Yang, L.; Zhang, D.; Shi, J. Plant metabolomics: An indispensable system biology tool for plant science. Int. J. Mol. Sci. 2016, 17, 767. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, N.M.; Bouwmeester, H.J. Metabolomics in the rhizosphere: Tapping into belowground chemical communication. Trends Plant Sci 2016, 21, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Erban, A.; Weber, R.J.; Creek, D.J.; Brown, M.; Breitling, R.; Hankemeier, T.; Goodacre, R.; Neumann, S.; Kopka, J.; et al. Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 2013, 9, 44–66. [Google Scholar] [CrossRef]
- Hardy, N.W.; Hall, R.D.E. Plant Metabolomics: Methods and Protocols; Springer: New York, NY, USA, 2012. [Google Scholar]
- Hoekenga, O.A.; Vision, T.J.; Shaff, J.E.; Monforte, A.J.; Lee, G.P.; Howell, S.H.; Kochian, L.V. Identification and characterization of aluminum tolerance loci in arabidopsis (Landsberg erecta). Plant Physiol. 2003, 132, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Han, X.; Zhu, X.; Herbert, S.J. Response to water stress of soil enzymes and root exudates from drought and non-drought tolerant corn hybrids at different growth stages. Can. J. Soil Sci. 2012, 92, 501–507. [Google Scholar] [CrossRef]
- Nimbal, C.I.; Pedersen, J.F.; Yerkes, C.N.; Weston, L.A.; Weller, S.C. Phytotoxicity and distribution of sorgoleone in grain sorghum germplasm. J. Agric. Food Chem. 1996, 44, 1343–1347. [Google Scholar] [CrossRef]
- Boeuf-Tremblay, V.; Plantureux, S.; Guckert, A. Influence of mechanical impedance on root exudation of maize seedlings at two development stages. Plant Soil 1995, 172, 279–287. [Google Scholar] [CrossRef]
- Watt, M.; McCully, M.E.; Kirkegaard, J.A. Soil strength and rate of root elongation alter the accumulation of pseudomonas spp. And other bacteria in the rhizosphere of wheat. Funct. Plant Biol. 2003, 30, 483–491. [Google Scholar] [CrossRef]
- Fernandez, O.; Béthencourt, L.; Quero, A.; Sangwan, R.S.; Clément, C. Trehalose and plant stress responses: Friend or foe? Trends Plant Sci. 2010, 15, 409–417. [Google Scholar] [CrossRef]
- Lopez-Bucio, J.; Nieto-Jacobo, M.F.; Ramırez-Rodrıguez, V.; Herrera-Estrella, L. Organic acid metabolism in plants: From adaptive physiology to transgenic varieties for cultivation in extreme soils. Plant Sci. 2000, 160, 1–13. [Google Scholar] [CrossRef]
- Jones, D.L. Organic acids in the rhizosphere—A critical review. Plant Soil 1998, 2015, 25–44. [Google Scholar] [CrossRef]
- Guo, J.; Carrington, Y.; Alber, A.; Ehlting, J. Molecular characterization of quinate and shikimate metabolism in populus trichocarpa. J. Biol. Chem. 2014, 289, 23846–23858. [Google Scholar] [CrossRef]
- Minamikawa, T. A comparative study on the metabolism of quinic and shikimic acids in plants. J. Plant Res. 1976, 89, 141–144. [Google Scholar] [CrossRef]
- Weston, L.A.; Mathesisus, U. Root exudation: The role of secondary metabolites, their localisation in roots and transport into the rhizosphere. In Root Engineering. Soil Biology; Morte, A., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 40, pp. 221–247. [Google Scholar]
- Hunter, P.J.; Teakle, G.R.; Bending, G.D. Root traits and microbial community interactions in relation to phosphorus availability and acquisition, with particular reference to brassica. Front. Plant Sci. 2014, 5, 27. [Google Scholar] [CrossRef]
- Rudrappa, T.; Czymmek, K.J.; Paré, P.W.; Bais, H.P. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008, 148, 1547–1556. [Google Scholar] [CrossRef] [PubMed]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behera, B.; Wagner, G.H. Microbial growth rate in glucose-amended soil. Soil Sci. Soc. Am. J. 1974, 38, 591–594. [Google Scholar] [CrossRef]
- Yang, Y.; Pollard, A.M.; Höfler, C.; Poschet, G.; Wirtz, M.; Hell, R.; Sourjik, V. Relation between chemotaxis and consumption of amino acids in bacteria. Mol. Microbiol. 2015, 96, 1272–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spaepen, S.; Vanderleyden, J. Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef] [PubMed]
- Barea, J.M.; Navarro, E.; Montoya, E. Production of plant growth regulators by rhizosphere phosphate-solubilizing bacteria. J. Appl. Microbiol. 1976, 40, 129–134. [Google Scholar] [CrossRef]
- Frankenberger, J.W.T.; Arshad, M. Microbial Synthesis of Auxins; Marcel Dekker: New York, NY, USA, 1995; pp. 35–71. [Google Scholar]
- Bolen, D.W. Protein Stabilization by Naturally Occurring Osmolytes. Methods Mol. Biol. 2001, 168, 17–36. [Google Scholar] [PubMed]
- Shen, B.; Hohmann, S.; Jensen, R.G.; Bohnert, H.J. Roles of sugar alcohols in osmotic stress adaptation. Replacement of glycerol by mannitol and sorbitol in yeast. Plant Physiol. 1999, 121, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Nikel, P.I.; Romero-Campero, F.J.; Zeidman, J.A.; Goñi-Moreno, Á.; de Lorenzo, V. The glycerol-dependent metabolic persistence of pseudomonas putida kt2440 reflects the regulatory logic of the glpr repressor. mBio 2015, 6, e00340-15. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhang, Y.; Wang, J.; Zhou, Y. Glycerol affects root development through regulation of multiple pathways in arabidopsis. PLoS ONE 2014, 9, e86269. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Si, P.; Shao, W.; Quiao, X.; Yang, X.; Gao, D.; Wang, Z. Response of enzyme activities and microbial communities to soil amendment with sugar alchohols. Microbiologyopen 2016, 5, 604–615. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Mönchgesang, S.; Strehmel, N.; Trutschel, D.; Westphal, L.; Neumann, S.; Scheel, D. Plant-to-plant variability in root metabolite profiles of 19 arabidopsis thaliana accessions as substance-slass-dependent. Int. J. Mol. Sci. 2016, 17, 1565. [Google Scholar] [CrossRef]
- Pétriacq, P.; Williams, A.; Cotton, T.E.; McFarlane, A.E.; Rolfe, S.A.; Ton, J. Metabolite profiling of non-sterile rhizosphere soil. Plant J. 2017, 92, 147–162. [Google Scholar] [CrossRef] [Green Version]
- Vinaixa, M.; Samino, S.; Saez, I.; Duran, J.; Guinovart, J.J.; Yanes, O. A guideline to univariate statistical analysis for lc/ms-based untargeted metabolomics-derived data. Metabolites 2012, 2, 775–795. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.J. The plant hormones: Their nature, occurrence, and functions. In Plant Hormones; Springer: Dordrecht, The Netherlands, 2010; pp. 1–15. [Google Scholar]
- Moe, L.A. Amino acids in the rhizosphere: From plants to microbes. Am. J. Bot. 2013, 100, 1692–1705. [Google Scholar] [CrossRef] [Green Version]
- Tzin, V.; Galili, G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 2010, 3, 956–972. [Google Scholar] [CrossRef] [PubMed]
- Busk, P.K.; Møller, B.L. Dhurrin synthesis in sorghum is regulated at the transcriptional level and induced by nitrogen fertilization in older plants. Plant Physiol. 2002, 129, 1222–1231. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; John, E.B.; Rémy, B.; Inna, D.; Jane, G.; Heidrun, G.; Georg, H.; Uffe, H.; Therese, M.; Alexander, P.; et al. The sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.H.; Bhaskaran, S.; Miller, F.R. Screening for drought tolerance in sorghum using cell culture. In Vitro Cell. Dev. Biol. Plant 1985, 21, 541–545. [Google Scholar] [CrossRef]
- Kebede, H.; Subudhi, P.K.; Rosenow, D.T.; Nguyen, H.T. Quantitative trait loci influencing drought tolerance in grain sorghum (Sorghum bicolor l. Moench). Theor. Appl. Genet. 2001, 103, 266–276. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Peredes, S.H.; Yourstone, S.; Gehriing, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. Xcms: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Broeckling, C.D.; Afsar, F.A.; Neumann, S.; Ben-Hur, A.; Prenni, J.E. Ramclust: A novel feature clustering method enables spectral-matching-based annotation for metabolomics data. Anal. Chem. 2014, 86, 6812–6817. [Google Scholar] [CrossRef]
- Broeckling, C.D.; Ganna, A.; Layer, M.; Brown, K.; Sutton, B.; Ingelsson, E.; Peers, G.; Prenni, J.E. Enabling efficient and confident annotation of lc-ms metabolomics data through ms1 spectrum and time prediction. Anal. Chem. 2016, 88, 9226–9234. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]





| Substrate | Total Metabolites of Interest | GC-MS | UPLC-MS | REMs | RAMs |
|---|---|---|---|---|---|
| Sand | 162 | 119 | 43 | 105 | 57 |
| Clay | 135 | 113 | 22 | 73 | 62 |
| Soil | 13 | 9 | 4 | 11 | 2 |
| Sand | Clay | Soil | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | Platform Detected | BTx623 | SC56 | BTx623 | SC56 | BTx623 | SC56 | ||||||
| Amino Acids | log2 FC | p-value | log2 FC | p-value | log2 FC | p-value | log2 FC | p-value | log2 FC | p-value | log2 FC | p-value | |
| alanine (2) | GC-MS | 4.28 | 0.0002 | 4.24 | 0.0820 | 3.21 | 0.0137 | 2.71 | 0.0150 | 1.29 | 0.0895 | 1.05 | 0.3161 |
| B-alanine (2) | GC-MS | 3.67 | 0.0012 | 3.97 | 0.0379 | 1.37 | 0.4369 | 1.96 | 0.0851 | 0.78 | 0.1206 | 0.69 | 0.6791 |
| aminobutyric acid (2) | GC-MS | 2.50 | 0.0026 | 2.51 | 0.0724 | 0.87 | 0.3710 | 1.25 | 0.1438 | 0.80 | 0.2942 | 0.85 | 0.6579 |
| glycine (2) | GC-MS | 2.04 | 0.0100 | 2.02 | 0.0617 | 0.54 | 0.7521 | 1.08 | 0.0631 | 0.49 | 0.3633 | 0.52 | 0.7180 |
| leucine (2) | UPLC-MS | 5.25 | 0.0001 | 5.59 | 0.0215 | 4.26 | 0.0258 | 3.90 | 0.0251 | 0.87 | 0.3550 | 0.51 | 0.7694 |
| phenylalanine (2) | UPLC-MS | 7.48 | 0.0256 | 7.34 | 0.0205 | 4.56 | 0.0134 | 4.73 | 0.0042 | 1.43 | 0.2979 | 1.46 | 0.2670 |
| pyroglutamate (2) | GC-MS | 3.45 | 0.2487 | 2.57 | 0.4107 | 3.36 | 0.4140 | 1.43 | 0.2219 | 1.01 | 0.5871 | 2.09 | 0.4284 |
| serine (2) | GC-MS | 4.02 | 0.0002 | 3.85 | 0.0219 | 3.59 | 0.0020 | 2.90 | 0.0261 | 1.99 | 0.1103 | 1.76 | 0.2145 |
| threonine (2) | GC-MS | 4.42 | 0.0010 | 4.90 | 0.0995 | 2.45 | 0.3479 | 3.69 | 0.0851 | 0.71 | 0.2389 | 0.55 | 0.7657 |
| tryptamine (2) | UPLC-MS | 4.97 | 0.1013 | 5.22 | 0.0213 | 5.68 | 0.0121 | 5.70 | 0.0113 | 1.25 | 0.0797 | 2.36 | 0.1350 |
| tryptophan (2) | UPLC-MS | 6.88 | 0.0033 | 7.76 | 0.0122 | 4.73 | 0.0469 | 6.27 | 0.0113 | 2.96 | 0.2371 | 3.70 | 0.1547 |
| tyrosine (2) | UPLC-MS | 6.12 | 0.0030 | 5.47 | 0.0280 | 4.54 | 0.0469 | 4.15 | 0.0127 | 2.76 | 0.0645 | 1.92 | 0.3013 |
| valine (2) | GC-MS | 4.33 | 0.0007 | 4.40 | 0.0811 | 2.76 | 0.3001 | 3.71 | 0.0241 | 1.33 | 0.0645 | 0.90 | 0.7087 |
| choline + glutamic acid (3) | UPLC-MS | 5.20 | 0.0065 | 5.04 | 0.0018 | 2.40 | 0.0145 | 2.19 | 0.0259 | 1.35 | 0.0645 | 1.06 | 0.1726 |
| C5H11NO2 (valine) (3) | UPLC-MS | 5.14 | 0.0008 | 2.45 | 0.0379 | 0.63 | 0.3200 | 0.53 | 0.2153 | −0.88 | 0.5546 | −1.59 | 0.4187 |
| Carbohydrates | |||||||||||||
| fructose (2) | GC-MS | 7.72 | 0.0008 | 7.14 | 0.0006 | 7.29 | 0.0034 | 6.80 | 0.0241 | 2.06 | 0.1461 | 2.18 | 0.2888 |
| glucose (2) | GC-MS | 8.09 | 0.0042 | 7.88 | 0.0067 | 6.65 | 0.0025 | 6.11 | 0.0247 | 0.24 | 0.2436 | 0.29 | 0.7694 |
| glycerol (2) | GC-MS | −2.84 | 0.0010 | −2.10 | 0.0015 | −1.48 | 0.0041 | −0.90 | 0.0261 | −0.49 | 0.7143 | −0.38 | 0.7425 |
| myo-inositol (2) | GC-MS | 4.60 | 0.0006 | 4.96 | 0.0004 | 4.39 | 0.0120 | 4.03 | 0.0113 | 0.31 | 0.2587 | 0.30 | 0.4533 |
| sucrose (2) | GC-MS | 6.53 | 0.1135 | 6.20 | 0.0666 | 8.56 | <0.0001 | 8.32 | 0.0200 | 5.21 | 0.0635 | 4.93 | 0.3333 |
| trehalose (2) | GC-MS | 3.95 | 0.2333 | 5.51 | 0.0489 | −1.46 | 0.2067 | 0.24 | 0.9132 | −0.33 | 0.9717 | 0.02 | 0.7694 |
| disaccharide 01 (3) | UPLC-MS | 6.01 | <0.0001 | 5.54 | 0.0101 | 3.04 | 0.0343 | 2.69 | 0.0289 | 0.49 | 0.5546 | 0.40 | 0.5050 |
| disaccharide 02 (3) | UPLC-MS | 6.90 | 0.0005 | 5.09 | 0.0080 | 1.23 | 0.0293 | 1.59 | 0.0560 | −0.49 | 0.6437 | −0.16 | 0.8950 |
| hexose sugar acid (3) | GC-MS | 1.10 | 0.0330 | 0.70 | 0.1017 | 4.13 | 0.0093 | 3.66 | 0.0627 | 1.15 | 0.1156 | 1.24 | 0.4533 |
| hexose + glutamine (3) | UPLC-MS | 5.93 | <0.0001 | 4.97 | 0.0010 | 3.48 | 0.0137 | 3.17 | 0.0259 | 0.94 | 0.1785 | 0.33 | 0.5323 |
| hexose 01 (3) | GC-MS | 7.54 | 0.0013 | 7.56 | 0.0026 | 6.24 | 0.0190 | 5.60 | 0.0498 | 0.05 | 0.2892 | 0.12 | 0.7679 |
| hexose 02 (3) | GC-MS | 4.39 | 0.0012 | 4.04 | 0.0051 | 3.93 | 0.0254 | 3.70 | 0.0368 | −0.08 | 0.7143 | 0.04 | 0.9964 |
| inositol-like (3) | GC-MS | 3.31 | 0.0508 | 2.84 | 0.0121 | 1.91 | 0.0237 | 1.41 | 0.0188 | −0.07 | 0.6744 | 0.10 | 0.8236 |
| pentose (3) | GC-MS | 3.67 | 0.0004 | 3.54 | 0.0158 | 3.74 | 0.0025 | 3.60 | 0.0244 | 1.05 | 0.1785 | 0.99 | 0.3661 |
| sugar alcohol 01 (3) | GC-MS | 5.16 | 0.1614 | 5.68 | 0.0486 | 3.25 | 0.5230 | 3.94 | 0.2763 | −0.13 | 0.8860 | 0.65 | 0.8556 |
| sugar alcohol 02 (3) | GC-MS | 5.94 | 0.2057 | 7.22 | 0.0382 | 0.19 | 0.9370 | 1.64 | 0.3052 | −.31 | 0.1206 | −1.75 | 0.0286 |
| sugar alcohol 03 (3) | GC-MS | 0.58 | 0.2693 | 0.37 | 0.2932 | 0.89 | 0.2100 | 0.64 | 0.0955 | 0.69 | 0.1557 | 2.01 | 0.2259 |
| trisaccharide (3) | GC-MS | −0.21 | 0.7571 | 0.07 | 0.9323 | −0.67 | 0.3383 | −0.58 | 0.3475 | 0.49 | 0.6368 | 2.01 | 0.4472 |
| Organic Acids | |||||||||||||
| aconitic acid (2) | GC-MS | 4.51 | 0.5198 | 1.70 | 0.0802 | 0.44 | 0.8463 | 0.54 | 0.7111 | 2.18 | 0.6733 | 1.74 | 0.4837 |
| glyceric acid (2) | GC-MS | 1.59 | 0.0198 | 0.99 | 0.1468 | 2.29 | 0.0818 | 1.50 | 0.0749 | 0.55 | 0.3271 | 0.77 | 0.4444 |
| malic acid (2) | GC-MS | 6.02 | 0.2159 | 6.25 | 0.1816 | 3.17 | 0.0559 | 2.43 | 0.0599 | 2.39 | 0.4720 | 2.07 | 0.2145 |
| quinic acid (2) | GC-MS | 5.11 | 0.0085 | 4.36 | 0.0136 | 4.40 | 0.0015 | 3.85 | 0.0276 | 3.53 | 0.0927 | 3.25 | 0.0320 |
| threonic acid (2) | GC-MS | 5.42 | 0.0053 | 5.68 | 0.0234 | 5.88 | 0.0015 | 5.82 | 0.0379 | 3.01 | 0.1206 | 2.68 | 0.1350 |
| Vitamins | |||||||||||||
| pantothenic acid (2) | UPLC-MS | 5.44 | 0.0269 | 4.72 | 0.0176 | 5.31 | 0.0249 | 4.49 | 0.0181 | 4.64 | 0.1206 | 3.78 | 0.1753 |
| Other | |||||||||||||
| dhurrin (2) | UPLC-MS | 8.27 | 0.0235 | 7.59 | 0.0214 | 7.33 | 0.0015 | 6.50 | <0.0001 | 5.55 | 0.0348 | 5.11 | 0.0286 |
| prolyl-histidine-like (3) | UPLC-MS | 2.41 | 0.2219 | 2.07 | 0.0529 | 8.23 | 0.0134 | 8.65 | 0.0045 | 0.84 | 0.0927 | 0.97 | 0.1753 |
| tyrosyl-histidine-like (3) | UPLC-MS | 7.98 | 0.0079 | 6.81 | 0.0200 | 6.77 | 0.0172 | 5.01 | 0.0149 | 5.49 | 0.0645 | 3.63 | 0.2259 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, S.B.; Heuberger, A.L.; Broeckling, C.D.; Jahn, C.E. Non-Targeted Metabolomics Reveals Sorghum Rhizosphere-Associated Exudates are Influenced by the Belowground Interaction of Substrate and Sorghum Genotype. Int. J. Mol. Sci. 2019, 20, 431. https://doi.org/10.3390/ijms20020431
Miller SB, Heuberger AL, Broeckling CD, Jahn CE. Non-Targeted Metabolomics Reveals Sorghum Rhizosphere-Associated Exudates are Influenced by the Belowground Interaction of Substrate and Sorghum Genotype. International Journal of Molecular Sciences. 2019; 20(2):431. https://doi.org/10.3390/ijms20020431
Chicago/Turabian StyleMiller, Sarah B., Adam L. Heuberger, Corey D. Broeckling, and Courtney E. Jahn. 2019. "Non-Targeted Metabolomics Reveals Sorghum Rhizosphere-Associated Exudates are Influenced by the Belowground Interaction of Substrate and Sorghum Genotype" International Journal of Molecular Sciences 20, no. 2: 431. https://doi.org/10.3390/ijms20020431
APA StyleMiller, S. B., Heuberger, A. L., Broeckling, C. D., & Jahn, C. E. (2019). Non-Targeted Metabolomics Reveals Sorghum Rhizosphere-Associated Exudates are Influenced by the Belowground Interaction of Substrate and Sorghum Genotype. International Journal of Molecular Sciences, 20(2), 431. https://doi.org/10.3390/ijms20020431