Exploration of the Effect of Blue Light on Functional Metabolite Accumulation in Longan Embryonic Calli via RNA Sequencing
Abstract
:1. Introduction
2. Results
2.1. Physiological and Biochemical Indexes of Longan ECs under Different Light Qualities
2.2. Basic Data Analysis of Transcriptome Sequencing
2.3. Differentially Expressed Genes in Longan ECs under Different Light Qualities
2.4. GO Enrichment Analysis of Differentially Expressed Genes in Longan ECs
2.5. KEGG Enrichment Analysis of Differentially Expressed Genes in Longan ECs
2.6. Primary and Secondary Metabolic Pathways in Longan ECs under Different Light Qualities
2.7. Longan EC Transcription Factors That Are Important for Photoresponsivity
2.8. Screening and Expression Analysis of Longan EC Blue Signal-Related Genes
3. Discussion
3.1. Cell Signaling Perception and Conduction in Longan ECs under the Blue Light Condition
3.2. Light Affects the Accumulation of Functional Metabolites in Longan ECs via the Ca2+ Signaling Pathway
3.3. Longan ECs Initiate ROS Clearance and DNA Repair Functions in Response to Blue Light
3.4. The Important Role of the Sulfur Metabolism Pathway in Longan EC Light-Affecting Functional Metabolites
3.5. Longan ECs Respond to Light via the TCA and PPP Pathways
3.6. Some Metabolic Pathways are Closely Related to the Synthesis of Biotin, Polysaccharides, Carotenoids, Alkaloids and Total Flavonoids in Longan ECs
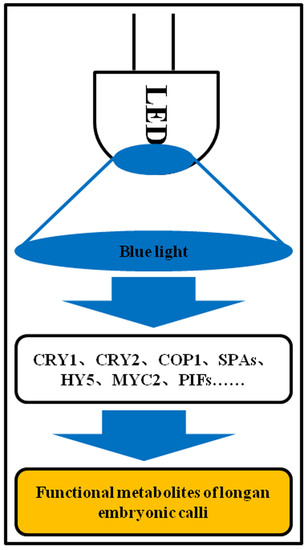
3.7. Blue light Signaling Network in Longan Functional Metabolites
4. Materials and Methods
4.1. Plant Material and Light Treatments
4.2. RNA-Seq Library Construction
4.3. Mapping Reads to the Reference Genome
4.4. Quantification of Gene Expression Levels
4.5. Differential Expression Analysis
4.6. GO and KEGG Enrichment Analyses of Differentially Expressed Genes
4.7. Carotenoid Determination
4.8. Measurement of Antioxidant Enzymes
4.9. Validation of the DEGs by qRT-PCR
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sudjaroen, Y.; Hull, W.E.; Erben, G.; Würtele, G.; Changbumrung, S.; Ulrich, C.M.; Owen, R.W. Isolation and characterization of ellagitannins as the major polyphenolic components of Longan (Dimocarpus longan Lour.) seeds. Phytochemistry 2012, 77, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Mao, W.; Han, Y.; Zhang, X.; Yang, C.; Chen, Y.; Chen, Y.; Xu, J.; Li, H.; Qi, X. Structural characteristics and antioxidant activities of the extracellular polysaccharides produced by marine bacterium Edwardsiella tarda. Bioresour. Technol. 2010, 101, 4729–4732. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Gao, L.P. Advances in biosynthesis pathways and regulation of flavonoids and catechins. Sci. Agric. Sin. 2009, 2899–2908. [Google Scholar]
- Li, W.; Guan, J.H.; Shi, L.L. Optimization of cell suspension culture system of Rhodiola fastigiata. Chin. Tradit. Herb. Drugs 2012, 43, 2272–2278. [Google Scholar]
- Dong, Y.L. Effects of Copper Ion Induction Elicitation on Camptothecin Biosynthesis in Cell Suspension Culture of Campototheca acuminata Decaisne. J. Anhui Agric. Sci. 2010, 38, 12457–12459. [Google Scholar]
- Zhang, Y.; Li, Y.; Zhou, J.; Chen, N.; Wang, M.; Dong, Z.; Gao, C.; Zhong, Y. Effect of precursor on growth and accumulation of alkaloids of Lycoris radiata suspension cells. Chin. J. Biotechnol. 2014, 30, 247–254. [Google Scholar]
- Lai, Z. Somatic embryogenesis of high frequency from longan embryogenic calli. J. Fujian Agric. Univ. 1997, 26, 271–276. [Google Scholar]
- Azuma, A.; Yakushiji, H.; Koshita, Y.; Kobayashi, S. Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta 2012, 236, 1067–1080. [Google Scholar] [CrossRef]
- Karppinen, K.; Zoratti, L.; Sarala, M.; Carvalho, E.; Hirsimäki, J.; Mentula, H.; Martens, S.; Häggman, H.; Jaakola, L. Carotenoid metabolism during bilberry (Vaccinium myrtillus L.) fruit development under different light conditions is regulated by biosynthesis and degradation. BMC Plant Biol. 2016, 16, 95. [Google Scholar] [CrossRef]
- Lai, B.; Li, X.J.; Hu, B.; Qin, Y.H.; Huang, X.M.; Wang, H.C.; Hu, G.B. LcMYB1 Is a Key Determinant of Differential Anthocyanin Accumulation among Genotypes, Tissues, Developmental Phases and ABA and Light Stimuli in Litchi chinensis. PLoS ONE 2014, 9, e86293. [Google Scholar] [CrossRef]
- Zhang, X.; Allan, A.C.; Yi, Q.; Chen, L.; Li, K.; Shu, Q.; Su, J. Differential Gene Expression Analysis of Yunnan Red Pear, Pyrus Pyrifolia, During Fruit Skin Coloration. Plant Mol. Biol. Report. 2011, 29, 305–314. [Google Scholar] [CrossRef]
- Linwang, K.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; Mcghie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 50. [Google Scholar]
- Koyama, K.; Ikeda, H.; Poudel, P.R.; Gotoyamamoto, N. Light quality affects flavonoid biosynthesis in young berries of Cabernet Sauvignon grape. Phytochemistry 2012, 78, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Lau, O.S.; Deng, X.W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007, 8, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Chen, Z.; Sun, W.; Deng, T.; Chen, M. De novo Sequencing of the Leaf Transcriptome Reveals Complex Light-Responsive Regulatory Networks in Camellia sinensis cv. Baijiguan. Front. Plant Sci. 2016, 7, 332. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Xu, Z.G.; Dong, R.Q.; Chang, S.X.; Wang, L.Z.; Khalilurrehman, M.; Tao, J.M. An RNA-Seq Analysis of Grape Plantlets Grown in vitro Reveals Different Responses to Blue, Green, Red LED Light and White Fluorescent Light. Front. Plant Sci. 2017, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Zhang, J.; Zhang, J.; Wang, Z.; An, R.; Guo, H.; Di, W.; Zhang, J. Integrated RNA-seq and sRNA-seq analysis reveals miRNA effects on secondary metabolism in Solanum tuberosum L. Mol. Genet. Genom. 2016, 292, 1–16. [Google Scholar]
- Xing, L.B.; Zhang, D.; Li, Y.M.; Shen, Y.W.; Zhao, C.P.; Ma, J.J.; An, N.; Han, M.Y. Transcription Profiles Reveal Sugar and Hormone Signaling Pathways Mediating Flower Induction in Apple (Malus domestica Borkh.). Plant Cell Physiol. 2015, 56, 2052. [Google Scholar] [CrossRef]
- Zhang, H.N.; Li, W.C.; Wang, H.C.; Shi, S.Y.; Bo, S.; Liu, L.Q.; Wei, Y.Z.; Xie, J.H. Transcriptome Profiling of Light-Regulated Anthocyanin Biosynthesis in the Pericarp of Litchi. Front. Plant Sci. 2016, 7, 963. [Google Scholar] [CrossRef]
- Ma, L.; Li, J.; Qu, L.; Hager, J.; Chen, Z.; Zhao, H.; Deng, X.W. Light Control of Arabidopsis Development Entails Coordinated Regulation of Genome Expression and Cellular Pathways. Plant Cell 2001, 13, 2589–2607. [Google Scholar] [CrossRef]
- Tepperman, J.M.; Zhu, T.; Chang, H.S.; Wang, X.; Quail, P.H. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 2001, 98, 9437–9442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Lin, Y.; Chen, X.; Bai, Y.; Wang, C.; Xu, X.; Wang, Y.; Lai, Z. Effects of blue light on flavonoid accumulation linked to the expression of miR393, miR394 and miR395 in longan embryogenic calli. PLoS ONE 2018, 13, e0191444. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chen, X.; Wang, Y.; Yao, D.; Lin, Y.; Lai, Z. Exploration of the effect of blue light on microRNAs involved in the accumulation of functional metabolites of longan embryonic calli through RNA-sequencing. J. Sci. Food Agric. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.S.; Chen, M.M.; Shi, P.U.; Liu, Y.; Feng, J.T. The Roles of Niacin, Aspartic Acid and Isolencine in Growth and Secondary Metabolites in Tripterygium wilfordii Hook. F. Suspension Cell. J. Xinjiang Agric. Univ. 2017, 40, 403–408. [Google Scholar]
- Kushalappa, A.C.; Raghavendra, G. Metabolo-proteomics to discover plant biotic stress resistance genes. Trends Plant Sci. 2013, 18, 522–531. [Google Scholar] [CrossRef]
- Docimo, T.; Francese, G.; Ruggiero, A.; Batelli, G.; De, P.M.; Bassolino, L.; Toppino, L.; Rotino, G.L.; Mennella, G.; Tucci, M. Phenylpropanoids Accumulation in Eggplant Fruit: Characterization of Biosynthetic Genes and Regulation by a MYB Transcription Factor. Front. Plant Sci. 2016, 6, 1233. [Google Scholar] [CrossRef]
- Filo, J.; Wu, A.; Eliason, E.; Richardson, T.; Thines, B.C.; Harmon, F.G. Gibberellin driven growth in elf3 mutants requires PIF4 and PIF5. Plant Signal. Behav. 2015, 10, e992707. [Google Scholar] [CrossRef]
- Guo, B.; Ying, L.I.; Yuan, Z.; Chao, L.; Zhang, X.; Rugen, X.U. Genome-Wide Analysis of Auxin Response Factor (ARF) Family in Barley. J. Triticeae Crops 2016, 36, 1426–1432. [Google Scholar]
- Kemmerling, B.; Schwedt, A.; Rodriguez, P.; Mazzotta, S.; Frank, M.; Qamar, S.A.; Mengiste, T.; Betsuyaku, S.; Parker, J.E.; Müssig, C. The BRI1-Associated Kinase 1, BAK1, Has a Brassinolide-Independent Role in Plant Cell-Death Control. Curr. Biol. 2007, 17, 1116–1122. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Liu, H.; Klejnot, J.; Lin, C. The Cryptochrome Blue Light Receptors. Arab. Book 2010, 8, e0135. [Google Scholar] [CrossRef]
- Wang, Q.; Barshop, W.D.; Bian, M.; Vashisht, A.A.; He, R.; Yu, X.; Liu, B.; Nguyen, P.; Liu, X.; Zhao, X.; et al. The Blue Light-Dependent Phosphorylation of the CCE Domain Determines the Photosensitivity of Arabidopsis CRY2. Mol. Plants 2017, 8, 357. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Li, H.; Wang, Q.; Liu, B.; Lin, C. Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell 2013, 25, 4405–4420. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Y.; Wang, J.; Li, P.; Zhao, C.; Chen, Y.; Bi, Y. Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis seedlings. Plant Sci. 2015, 238, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, B.; Su, J.; Liao, J.; Lin, C.; Oka, Y. Cryptochromes Orchestrate Transcription Regulation of Diverse Blue Light Responses in Plants. Photochem. Photobiol. 2017, 93, 112–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Jiang, J.; Ren, X.; Li, L.; Li, X.; Zhao, H. Effect of Low Temperature Stress on Cell Membrane and Osmoregulatory Substance in Three Varieties of Citrus reticulate Blanco Seedlings. Mol. Plant Breed. 2017, 15, 1104–1108. [Google Scholar]
- Graier, W.F.; Paltauf-Doburzynska, J.; Hill, B.J.F.; Fleischhacker, E.; Hoebel, B.G.; Kostner, G.M.; Sturek, M. Submaximal stimulation of porcine endothelial cells causes focal Ca2+ elevation beneath the cell membrane. J. Physiol. 2010, 506, 109–125. [Google Scholar] [CrossRef]
- Abdullahi, A.S.; Nzelibe, H.C.; Atawodi, S.E. Purification, Characterization and Vaccine Potential of Glycosyl Phosphatidyl Inositol Specific Phospholipase C. Int. J. Trop. Dis. Health 2015, 5, 71–83. [Google Scholar] [CrossRef]
- Li, H.; Zhou, H.; Luo, Y.; Ouyang, H.; Hu, H.; Cheng, J. Glycosylphosphatidylinositol (GPI) anchor is required in Aspergillus fumigatus for morphogenesis and virulence. Mol. Microbiol. 2010, 64, 1014–1027. [Google Scholar] [CrossRef] [PubMed]
- Yeats, T.H.; Bacic, A.; Johnson, K.L. Plant glycosylphosphatidylinositol (GPI) anchored proteins at the plasma membrane-cell wall nexus. J. Integr. Plant Biol. 2018, 60, 649–669. [Google Scholar] [CrossRef] [PubMed]
- Immonen, E.V.; Krause, S.; Krause, Y.; Frolov, R.; Vähäsöyrinki, M.T.; Weckström, M. Elementary and macroscopic light-induced currents and their Ca2+-dependence in the photoreceptors of Periplaneta americana. Front. Physiol. 2014, 5, 153. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.W.; Zhou, W. Effects of calcium on content of calmodulin, activity of Ca-ATPase and their gene expressions in apple (Mill.) fruits. Plant Nutr. Fertil. Sci. 2011, 17, 425–432. [Google Scholar]
- Yang, M.Z.; Ye, W.U.; Zhang, Y.D. The Effect of the CAM and CAM-Dependent Kinase on the Expression of c-fos Gene in the Spinal Cord Matter. J. Nanhua Univ. 2007, 35, 529–532. [Google Scholar]
- Živanović, B.D.; Shabala, L.I.; Elzenga, T.J.M.; Shabala, S.N. Dissecting blue light signal transduction pathway in leaf epidermis using a pharmacological approach. Planta 2015, 242, 813–827. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.T.; Zhang, J.W.; Wang, D.; Tao, S.H.; Ji, Y.L.; Wu, B. Relation between light qualities and accumulation of steroidal glycoalkaloids as well as signal molecule in cell in potato tubers. Acta Agron. Sin. 2010, 36, 629–635. [Google Scholar] [CrossRef]
- Cao, R.R.; Dang, X.L.; Xing, B.Y.; Zhang, J.Y.; Dong, J.E. Regulation effects of intracellular and extracellular Ca2+ on biosynthesis of rosmarinic acid induced by salicylic acid in young seedlings of Salvia miltiorrhiza. Zhongguo Zhong Yao Za Zhi 2013, 38, 3424–3431. [Google Scholar] [PubMed]
- Lushchak, V.I. Adaptive response to oxidative stress: Bacteria, fungi, plants and animals. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011, 153, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Shulaev, V.; Mittler, R. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 2010, 133, 481–489. [Google Scholar] [CrossRef]
- Luo, Y.L.; Song, S.Q.; Lan, Q.Y. Possible involvement of enzymatic and non-enzymatic antioxidant system in acquisition of desiccation tolerance of maize embryos. Acta Bot. Yunnanica 2009, 31, 253–259. [Google Scholar] [CrossRef]
- Algubory, K.H.; Fowler, P.A.; Garrel, C. The roles of cellular reactive oxygen species, oxidative stress and antioxidants in pregnancy outcomes. Int. J. Biochem. Cell Biol. 2010, 42, 1634–1650. [Google Scholar] [CrossRef]
- Ravilious, G.E.; Jez, J.M. Structural biology of plant sulfur metabolism: From assimilation to biosynthesis. Nat. Prod. Rep. 2012, 29, 1138–1152. [Google Scholar] [CrossRef]
- Hubberten, H.M.; Klie, S.; Caldana, C.; Degenkolbe, T.; Willmitzer, L.; Hoefgen, R. Additional role of O-acetylserine as a sulfur status-independent regulator during plant growth. Plant J. 2012, 70, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Brychkova, G.; Xia, Z.; Yang, G.; Yesbergenova, Z.; Zhang, Z.; Davydov, O.; Fluhr, R.; Sagi, M. Sulfite oxidase protects plants against sulfur dioxide toxicity. Plant J. 2010, 50, 696–709. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.J.; Zhang, S.L.; Li, D.F.; Zhao, Y.Z.; Tian, X.L.; Zhao, X.L.; Wu, Y.X.; Wei, X.Y.; Liu, R.Q. Effects of exogenous hydrogen sulfide on the ascorbate and glutathione metabolism in wheat seedlings leaves under water stress. Acta Physiol. Plant. 2011, 33, 2533–2540. [Google Scholar] [CrossRef]
- Capaldi, F.R.; Gratão, P.L.; Reis, A.R.; Lima, L.W.; Azevedo, R.A. Sulfur Metabolism and Stress Defense Responses in Plants. Trop. Plant Biol. 2015, 8, 60–73. [Google Scholar] [CrossRef]
- Mugford, S.G.; Lee, B.R.; Koprivova, A.; Matthewman, C.; Kopriva, S. Control of sulfur partitioning between primary and secondary metabolism. Plant J. 2011, 65, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Che, F.; Dong, C.; Zhang, H.; Meng, X.; Zhang, T. Effects of simulated transport vibration on respiratory pathways and qualities of Xinjiang apricot fruit. Trans. Chin. Soc. Agric. Eng. 2015, 31, 325–331. [Google Scholar]
- Nunes-Nesi, A.; Sweetlove, L.J.; Fernie, A.R. Operation and function of the tricarboxylic acid cycle in the illuminated leaf. Physiol. Plant. 2010, 129, 45–56. [Google Scholar] [CrossRef]
- Marrero, J.; Rhee, K.Y.; Schnappinger, D.; Pethe, K.; Ehrt, S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. USA 2010, 107, 9819–9824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronimus, R.S.; Morgan, H.W. Distribution and phylogenies of enzymes of the Embden-Meyerhof-Parnas pathway from archaea and hyperthermophilic bacteria support a gluconeogenic origin of metabolism. Archaea 2003, 1, 199. [Google Scholar] [CrossRef] [PubMed]
- Komati, R.G.; Lindner, S.N.; Wendisch, V.F. Metabolic engineering of an ATP-neutral Embden-Meyerhof-Parnas pathway in Corynebacterium glutamicum: Growth restoration by an adaptive point mutation in NADH dehydrogenase. Appl. Environ. Microbiol. 2015, 81, 1996–2005. [Google Scholar] [CrossRef]
- Nakanishi, S.; Numa, S. Purification of Rat Liver Acetyl Coenzyme A Carboxylase and Immunochemical Studies on its Synthesis and Degradation. Eur. J. Biochem. 2010, 16, 161–173. [Google Scholar] [CrossRef]
- Liu, L.J.; Zhang, Y.C.; Li, Q.H.; Sang, Y.; Mao, J.; Lian, H.L.; Wang, L.; Yang, H.Q. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 2008, 20, 292–306. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 Interacts with CIB1 to Regulate Transcription and Floral Initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Li, X.; Guo, Y.; Chu, J.; Fang, S.; Yan, C.; Noel, J.P.; Liu, H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 2016, 113, 224. [Google Scholar] [CrossRef] [PubMed]
- Pedmale, U.V.; Huang, S.S.; Zander, M.; Cole, B.J.; Hetzel, J.; Ljung, K.; Reis, P.A.; Sridevi, P.; Nito, K.; Nery, J.R. Cryptochromes Interact Directly with PIFs to Control Plant Growth in Limiting Blue Light. Cell 2016, 164, 233–245. [Google Scholar] [CrossRef] [PubMed]
- De, W.M.; Keuskamp, D.H.; Bongers, F.J.; Hornitschek, P.; Gommers, C.M.; Reinen, E.; Martã Nez-Cerã, N.C.; Fankhauser, C.; Pierik, R. Integration of Phytochrome and Cryptochrome Signals Determines Plant Growth during Competition for Light. Curr. Biol. 2016, 26, 3320–3326. [Google Scholar]
- Opseth, L.; Holefors, A.; Rosnes, A.K.R.; Lee, Y.K.; Olsen, J.E. FTL2 expression preceding bud set corresponds with timing of bud set in Norway spruce under different light quality treatments. Environ. Exp. Bot. 2015, 204, 121–131. [Google Scholar] [CrossRef]
- Zhu, T.; Lin, L.; Yang, T.; Gao, F.; Liu, T.; Pan, D.; Xia, H. Progress in Anthocyanin Biosynthesis of Plants Regulated by COP1. Mol. Plant Breed. 2015, 13, 2135–2140. [Google Scholar]
- Yu, J.W.; Rubio, V.; Lee, N.Y.; Bai, S.; Lee, S.Y.; Kim, S.S.; Liu, L.; Zhang, Y.; Irigoyen, M.L.; Sullivan, J.A. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 2008, 32, 617–630. [Google Scholar] [CrossRef]
- Hong, S.H.; Kim, H.J.; Ryu, J.S.; Choi, H.; Jeong, S.; Shin, J.; Choi, G.; Hong, G.N. CRY1 inhibits COP1-mediated degradation of BIT1, a MYB transcription factor, to activate blue light-dependent gene expression in Arabidopsis. Plant J. 2010, 55, 361–371. [Google Scholar] [CrossRef]
- Li, Y.Y.; Mao, K.; Zhao, C.; Zhao, X.Y.; Zhang, H.L.; Shu, H.R.; Hao, Y.J. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 2012, 160, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ouyang, X.; Deng, X.W. Beyond repression of photomorphogenesis: Role switching of COP/DET/FUS in light signaling. Curr. Opin. Plant Biol. 2014, 21, 96–103. [Google Scholar] [CrossRef]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef]
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Horticul. Res. 2017, 4, 17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurya, J.P.; Sethi, V.; Gangappa, S.N.; Gupta, N.; Chattopadhyay, S. Interaction of MYC2 and GBF1 results in functional antagonism in blue light-mediated Arabidopsis seedling development. Plant J. Cell Mol. Biol. 2015, 83, 439–450. [Google Scholar] [CrossRef]
- Zhang, H.; Hedhili, S.; Montiel, G.; Zhang, Y.; Chatel, G.; Pré, M.; Gantet, P.; Memelink, J. The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. Cell Mol. Biol. 2011, 67, 61–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangappa, S.N.; Maurya, J.P.; Yadav, V.; Chattopadhyay, S. The Regulation of the Z- and G-Box Containing Promoters by Light Signaling Components, SPA1 and MYC2, in Arabidopsis. PLoS ONE 2013, 8, e62194. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Liu, R.; Hao, H.; Wang, Z.; Bi, Y. Phytochrome interacting factors (PIFs) are essential regulators for sucrose-induced hypocotyl elongation in Arabidopsis. J. Plant Physiol. 2011, 168, 1771. [Google Scholar] [CrossRef]
- Moon, J.; Zhu, L.; Shen, H.; Huq, E. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 9433–9438. [Google Scholar] [CrossRef] [Green Version]
- Stephenson, P.G.; Fankhauser, C.; Terry, M.J.; Lagarias, J.C. PIF3 Is a Repressor of Chloroplast Development. Proc. Natl. Acad. Sci. USA 2009, 106, 7654–7659. [Google Scholar] [CrossRef]
- Shin, J.; Park, E.; Choi, G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007, 49, 981–994. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, F.; Mao, J.F.; Wang, J.; Zhang, S.; Li, Y. Transcriptome Analysis Reveals that Red and Blue Light Regulate Growth and Phytohormone Metabolism in Norway Spruce [Picea abies (L.) Karst.]. PLoS ONE 2015, 10, e0127896. [Google Scholar] [CrossRef]
- Lin, Y.; Min, J.; Lai, R.; Wu, Z.; Chen, Y.; Yu, L.; Cheng, C.; Jin, Y.; Tian, Q.; Liu, Q. Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. Gigascience 2017, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.H.; Huang, G.W.; Zhu, Z.Y.; Jun-Yan, L.I.; Deng, J. Study on the extraction and stability of carotenoid in pomelo peel. Sci. Technol. Food Ind. 2014, 35, 281–285. [Google Scholar]
- Lin, Y.L.; Lai, Z.X. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci. 2010, 178, 359–365. [Google Scholar] [CrossRef]










| Sample | Dark | Blue | White |
|---|---|---|---|
| Total Raw Reads (Mb) | 74.35 | 74.35 | 74.35 |
| Total Clean Reads | 66,451,578 | 65,308,954 | 65,116,948 |
| Total Clean Bases (Gb) | 6.65 | 6.53 | 6.51 |
| Q20 (%) | 99.20 | 99.26 | 99.28 |
| Q30 (%) | 97.26 | 97.43 | 97.47 |
| Total Mapping Ratio (%) | 86.90 | 87.41 | 87.86 |
| Uniquely Mapping Ratio (%) | 70.30 | 71.21 | 71.55 |
| Pathway Term | Group | |||
|---|---|---|---|---|
| 1 | Nicotinate and nicotinamide metabolism | DB | DW | WB |
| 2 | β-Alanine metabolism | DB | DW | WB |
| 3 | RNA polymerase | DB | DW | |
| 4 | Protein processing in endoplasmic reticulum | DB | DW | |
| 5 | α-Linolenic acid metabolism | DB | DW | |
| 6 | Nucleotide excision repair | DB | DW | |
| 7 | Zeatin biosynthesis | DB | DW | |
| 8 | Galactose metabolism | DB | WB | |
| 9 | Ascorbate and aldarate metabolism | DB | WB | |
| 10 | Fatty acid biosynthesis | DB | WB | |
| 11 | Pyrimidine metabolism | DB | WB | |
| 12 | Lysine biosynthesis | DB | WB | |
| 13 | Biotin metabolism | DB | WB | |
| 14 | Nonhomologous end-joining | DB | WB | |
| 15 | RNA degradation | DB | WB | |
| 16 | Valine, leucine and isoleucine degradation | DB | WB | |
| 17 | Caffeine metabolism | DW | WB | |
| 18 | Taurine and hypotaurine metabolism | DB | DW | |
| 19 | Propanoate metabolism | DB | ||
| 20 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | DB | ||
| 21 | Phagosome | DB | ||
| 22 | Stilbenoid, diarylheptanoid and gingerol biosynthesis | DW | ||
| 23 | Plant hormone signal transduction | DW | ||
| 24 | Limonene and pinene degradation | DW | ||
| 25 | Glycerophospholipid metabolism | DW | ||
| 26 | N-Glycan biosynthesis | DW | ||
| 27 | Vitamin B6 metabolism | DW | ||
| 28 | Regulation of autophagy | DW | ||
| 29 | Brassinosteroid biosynthesis | DW | ||
| 30 | Alanine, aspartate and glutamate metabolism | DW | ||
| 31 | Other types of O-glycan biosynthesis | DW | ||
| 32 | Pyruvate metabolism | DW | ||
| 33 | Biosynthesis of unsaturated fatty acids | DW | ||
| 34 | Metabolic pathways | WB | ||
| 35 | Biosynthesis of secondary metabolites | WB | ||
| 36 | Fructose and mannose metabolism | WB | ||
| 37 | Carbon fixation in photosynthetic organisms | WB | ||
| 38 | Sulfur metabolism | WB | ||
| 39 | Pentose phosphate pathway | WB | ||
| 40 | Glycolysis/Gluconeogenesis | WB | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Lyu, Y.; Chen, X.; Wang, C.; Yao, D.; Ni, S.; Lin, Y.; Chen, Y.; Zhang, Z.; Lai, Z. Exploration of the Effect of Blue Light on Functional Metabolite Accumulation in Longan Embryonic Calli via RNA Sequencing. Int. J. Mol. Sci. 2019, 20, 441. https://doi.org/10.3390/ijms20020441
Li H, Lyu Y, Chen X, Wang C, Yao D, Ni S, Lin Y, Chen Y, Zhang Z, Lai Z. Exploration of the Effect of Blue Light on Functional Metabolite Accumulation in Longan Embryonic Calli via RNA Sequencing. International Journal of Molecular Sciences. 2019; 20(2):441. https://doi.org/10.3390/ijms20020441
Chicago/Turabian StyleLi, Hansheng, Yumeng Lyu, Xiaohui Chen, Congqiao Wang, Deheng Yao, Shanshan Ni, Yuling Lin, Yukun Chen, Zihao Zhang, and Zhongxiong Lai. 2019. "Exploration of the Effect of Blue Light on Functional Metabolite Accumulation in Longan Embryonic Calli via RNA Sequencing" International Journal of Molecular Sciences 20, no. 2: 441. https://doi.org/10.3390/ijms20020441
APA StyleLi, H., Lyu, Y., Chen, X., Wang, C., Yao, D., Ni, S., Lin, Y., Chen, Y., Zhang, Z., & Lai, Z. (2019). Exploration of the Effect of Blue Light on Functional Metabolite Accumulation in Longan Embryonic Calli via RNA Sequencing. International Journal of Molecular Sciences, 20(2), 441. https://doi.org/10.3390/ijms20020441