Differential Expression of Neuroinflammatory mRNAs in the Rat Sciatic Nerve Following Chronic Constriction Injury and Pain-Relieving Nanoemulsion NSAID Delivery to Infiltrating Macrophages
Abstract
:1. Introduction
2. Results
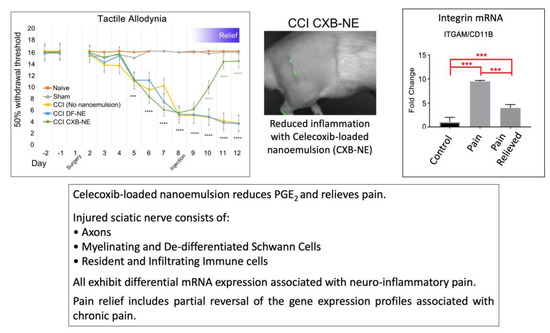
2.1. Relief of Pain Hypersensitivity in the CCI Model with a Drug-Loaded Nanoemulsion
2.2. Overview of mRNA Expression Changes for Genes Associated with CCI Chronic Pain
2.3. Altered Expression of Key Mrnas Involved in Neuroinflammation
2.4. Differential Expression of mRNAs in the CCI (Pain State) Compared to Celecoxib-Treated CCI (Pain Relieved)
2.5. Identification of CD68 and CD11b Macrophages within the Site of Injury
3. Discussion
3.1. Peripheral Nerve Injury is Associated with Differential Expression of Neuroinflammatory mRNAs
3.2. Targeting COX-2 in Macrophages Contributes to a Decrease in Macrophage-Related mRNAs at the Site of Injury
3.3. COX-2 Inhibition in Macrophages Leads to a Reduction in Pro-Inflammatory Cytokine mRNA
3.4. mRNAs are Differentially Expressed in Pain Relief versus the Untreated Pain State
3.5. mRNAs at the Site of Injury Contributing to Anti-Inflammation and Regeneration
3.6. Neuroinflammatory Genes Previously Associated with Cell Bodies Are Differentially Expressed at the Site of Injury
4. Materials and Methods
4.1. Animals
4.2. Peripheral Nerve Injury
4.3. Behavioral Testing
4.4. Nanoemulsion Preparation and Delivery
4.5. NIRF Imaging
4.6. Tissue Dissection
4.7. RNA Extraction and cDNA Conversion
4.8. Quantitative PCR and Analysis
4.9. Immunohistochemistry and Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CCI | Chronic constriction injury |
| NIRF | Near infrared fluorescence |
| CXB-NE | Celecoxib-loaded nanoemulsion |
| DF-NE | Drug-free nanoemulsion (vehicle) |
References
- Gaskin, D.J.; Richard, P. The economic costs of pain in the United States. J. Pain 2012, 13, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Bennett, G.J.; Xi, J. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain 1988, 33, 87–107. [Google Scholar] [CrossRef]
- Vasudeva, K.; Anderson, K.; Zeyzus-Johns, B.; Hitchens, T.K.; Patel, S.K.; Balducci, A.; Janjic, J.M.; Pollock, J.A. Imaging neuroinflammation in vivo in a neuropathic pain rat model with near-infrared fluorescence and 19F magnetic resonance. PLoS ONE 2014, 9, e90589. [Google Scholar] [CrossRef] [PubMed]
- Janjic, J.M.; Vasudeva, K.; Saleem, M.; Stevens, A.; Liu, L.; Patel, S.; Pollock, J.A. Low-dose NSAIDs reduce pain via macrophage targeted nanoemulsion delivery to neuroinflammation of the sciatic nerve in rat. J. Neuroimm. 2018, 318, 72–79. [Google Scholar] [CrossRef]
- Saleem, M.; Deal, B.; Nehl, E.; Janjic, J.M.; Pollock, J.A. Nanomedicine-driven neuropathic pain relief in a rat model is associated with macrophage polarity and mast cell activation. Acta. Neuropath. Comm. 2019, 7, 108. [Google Scholar] [CrossRef]
- DeLeo, J.; Yezierski, R.P. The role of neuroinflammation and neuroimmune activation in persistent pain. Pain 2001, 90, 1–6. [Google Scholar] [CrossRef]
- Grace, P.M.; Hutchinson, M.R.; Maier, S.F.; Watkins, L.R. Pathological pain and the neuroimmune interface. Nat. Rev. Imm. 2014, 14, 217–231. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.; van Niekerk, E.A.; Merianda, T.T.; Twiss, J.L. Dynamics of axonal mRNA transport and implications for peripheral nerve regeneration. Exp. Neur. 2010, 223, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Huebner, E.A.; Strittmatter, S.M. Axon regeneration in the peripheral and central nervous systems. Res. Prob. Cell Diff. 2009, 48, 339–351. [Google Scholar]
- Cattin, A.; Lloyd, A.C. The multicellular complexity of peripheral nerve regeneration. Curr. Opin. Neurobio. 2016, 39, 38–46. [Google Scholar] [CrossRef]
- Perry, V.H.; Tsao, J.W.; Fearn, S.; Brown, M.C. Radiation-induced reductions in macrophage recruitment have only slight effects on myelin degeneration in sectioned peripheral nerves of mice. Eur. J. Neurosci. 1995, 7, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Bruck, W. The role of macrophages in Wallerian degeneration. Brain Path. 1997, 7, 741–752. [Google Scholar] [CrossRef]
- Gaudet, A.D.; Popovich, P.G.; Ramer, M.S. Wallerian degeneration: Gaining perspective on inflammatory events after peripheral nerve injury. J. Neuroinfl. 2011, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Gu, N.; Zhou, L.; Evo, U.B.; Murugan, M.; Gan, W.; Wu, L. Microglia and monocytes synergistically promote the transition from acute to chronic pain after nerve injury. Nat. Comm. 2016, 7, e12029. [Google Scholar] [CrossRef]
- Costigan, M.; Scholz, J.; Woolf, C.J. Neuropathic pain: A maladaptive response of the nervous system to damage. Ann. Rev. Neurosci. 2009, 32, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.G.; Holmin, S.; Mathiesen, T.; Meyerson, B.A.; Linderoth, B. Possible role of inflammatory mediators in tactile hypersensitivity in rat models of mononeuropathy. Pain 2000, 88, 239–248. [Google Scholar] [CrossRef]
- Liu, T.; van Rooijen, N.; Tracey, D.J. Depletion of macrophages reduces axonal degradation and hyperalgesia following nerve injury. Pain 2000, 86, 25–32. [Google Scholar] [CrossRef]
- Ma, W.; Quirion, R. Up-regulation of interleukin-6 induced by prostaglandin E2 from invading macrophages following nerve injury: An in vivo and in vitro study. J. Neurochem. 2005, 93, 664–673. [Google Scholar] [CrossRef]
- Ristoiu, V. Contribution of macrophages to peripheral neuropathic pain pathogenesis. Life Sci. 2013, 93, 870–881. [Google Scholar] [CrossRef]
- Ginhoux, F.; Jung, S. Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat. Imm. Rev. 2014, 14, 392–404. [Google Scholar] [CrossRef]
- Stoll, G.; Griffin, J.W.; Li, C.Y.; Trapp, B.D. Wallerian degeneration in the peripheral nervous system: Participation of both Schwann cells and macrophages in myelin degradation. J. Neurocyt. 1989, 18, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, H.; Matsubara, T.; Mima, A.; Sumi, E.; Nagai, K.; Takahashi, T.; Abe, H.; Iehara, N.; Fukatsu, A.; Okamoto, H.; et al. Inhibition of MCP-1/CCR2 pathway ameliorates the development of diabetic neuropathy. Biomed. Biophys. Res. Comm. 2007, 360, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Maiguel, D.; Farid, M.H.; Wie, C.; Kuwano, Y.; Balla, K.M.; Hernandez, D.; Barth, C.J.; Lugo, G.; Donnelly, M.; Nayer, A.; et al. Small molecule-mediated activation of the integrin CD11b/CD18 reduces inflammatory disease. Sci. Signal. 2011, 4, ra57. [Google Scholar] [CrossRef] [PubMed]
- Heneka, M.T.; Kummer, M.P.; Latz, E. Innate immune activation in neurodegenerative disease. Nat. Rev. Imm 2014, 14, 463–477. [Google Scholar] [CrossRef]
- Parkitna, J.R.; Korostynski, M.; Kaminska-Chowaniec, D.; Obara, I.; Mika, J.; Przewlocka, B.; Przewlocki, R. Comparison of gene expression profiles in neuropathic and inflammatory pain. J. Phys. Pharm. 2006, 57, 401–414. [Google Scholar]
- Scholz, J.; Woolf, C.J. The neuropathic pain triad: Neurons, immune cells and glia. Nat. Neuro. 2007, 10, 1361–1368. [Google Scholar] [CrossRef]
- Austin, P.J.; Moalem-Taylor, G. The neuro-immune balance in neuropathic pain: Involvement of inflammatory immune cells, immune-like glial cells and cytokines. J. Neuroimm. 2010, 229, 26–50. [Google Scholar] [CrossRef]
- Dubovy, P.; Klusakova, I.; Svizenska, I.H. Inflammatory profiling of schwann cells in contact with growing axons distal to nerve injury. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
- Michaelis, M.; Vogel, C.; Blenk, K.H.; Arnarson, A.; Janig, W. Inflammatory mediators sensitize acutely axotomized nerve fibers to mechanical stimulation in the rat. J. Neurosci. 1998, 18, 7581–7587. [Google Scholar] [CrossRef]
- Patel, S.K.; Zhang, Y.; Pollock, J.A.; Janjic, J.M. Cycloxygenase-2 inhibiting perfluoropoly (ethylene glycol) ether theranostic nanoemulsions–in vitro study. PLoS ONE 2013, 8, e55802. [Google Scholar] [CrossRef]
- Patel, S.K.; Janjic, J.M. Macrophage targeted theranostics as personalized nanomedicine strategies for inflammatory disease. Theranostics 2015, 5, 150–172. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.K.; Beaino, W.; Andersen, C.J.; Janjic, J.M. Theranostic nanoemulsions for macrophage COX-2 inhibition in a murine inflammation model. Clin. Immun. 2015, 160, 59–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dixon, W.J. The up-and-down method for small samples. J. Am. Stat. Assn. 1965, 60, 967–978. [Google Scholar] [CrossRef]
- Chaplan, S.R.; Bach, F.W.; Pogrel, J.W.; Chung, J.M.; Yaksh, T.L. Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Meth. 1994, 53, 55–63. [Google Scholar] [CrossRef]
- Saleem, M.; Stevens, A.M.; Deal, B.; Liu, L.; Janjic, J.; Pollock, J.A. A new best practice for validating tail vein injections in rat with near-infrared-labeled agents. JoVE 2019, 146, e59295. [Google Scholar] [CrossRef]
- Janjic, J.M.; Patel, S.K.; Patrick, M.J.; Pollock, J.A.; DiVito, E.; Cascio, M. Suppressing inflammation from inside out with novel NIR visible perfluorocarbon nanotheranostics. Proc. SPIE 2013, 8596, 85960L. [Google Scholar]
- Janjic, J.M.; Shaw, P.; Zhang, S.; Yang, X.; Patel, S.K.; Bai, M. Perfluorocarbon nanoemulsions with fluorescent, colloidal and magnetic properties. Biomaterials 2014, 35, 4958–4968. [Google Scholar] [CrossRef] [Green Version]
- Vasudeva, K.; Vodovotz, Y.; Azhar, N.; Barclay, D.; Janjic, J.M.; Pollock, J.A. In vivo and systems biology studies implicate IL-18 as a central mediator in chronic pain. J. Neuroimm. 2015, 283, 43–49. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Zu, F.; Tang, Z.; Zhong, T.; Cao, J.; Guo, Q.; Huang, C. Distinct calcitonin gene-related peptide expression in primary afferents contribute to different neuropathic symptoms following chronic constriction or crush injuries to the rat sciatic nerve. Molec. Pain 2016, 12, 1–17. [Google Scholar] [CrossRef]
- Alexanian, A.; Sorokin, A. Cyclooxygenase 2: protein-protein interactions and posttranslational modifications. Physiol. Genom. 2017, 49, 667–681. [Google Scholar] [CrossRef]
- Cheever, T.R.; Olson, E.A.; Ervasti, J.M. Axonal regeneration and neuronal function are preserved in motor neurons lacking β-actin in vivo. PLoS ONE 2011, 6, e17768. [Google Scholar] [CrossRef] [PubMed]
- Hur, E.; Zhou, F. Growing the growth cone: Remodeling the cytoskeleton to promote axon regeneration. Trends Neurosci. 2012, 35, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Saegusa, H.; Kurihara, T.; Zong, S.; Kazuno, A.; Matsuda, Y.; Nonaka, T.; Han, W.; Toriyama, H.; Tanabe, T. Suppression of inflammatory and neuropathic pain symptoms in mice lacking the N-type Ca2+ channel. EMBO 2001, 20, 2349–2356. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Takami, K.; Kawai, Y.; Girgis, S.; Hillyard, C.J.; MacIntyre, I.; Emson, P.C. Distribution of calcitonin gene-related peptide in the rat peripheral nervous system with reference to its coexistence with substance P. Neurosci 1985, 15, 1227–1237. [Google Scholar] [CrossRef]
- Vause, C.V.; Durham, P.L. Calcitonin gene-related peptide differentially regulates gene and protein expression in trigeminal glia cells: Findings from array analysis. Neurosci. Lett. 2010, 473, 163–167. [Google Scholar] [CrossRef] [Green Version]
- Andersen, S.; Skorpen, F. Variation in the COMT gene: Implications for pain perception and pain treatment. Pharmacogenomics 2009, 10, 669–684. [Google Scholar] [CrossRef]
- Hartung, J.E.; Ciszek, B.P.; Nackley, A.G. Beta-2- and beta-3-adrenergic receptors drive COMT-dependent pain by increasing production of nitric oxide and cytokines. Pain 2014, 155, 1346–1355. [Google Scholar] [CrossRef]
- Pomonis, J.D.; Rogers, S.D.; Peteres, C.M.; Ghilardi, J.R.; Mantyh, P.W. Expression and localization of endothelin receptors: Implications for the involvement of peripheral glial in nociception. J. Neurosci. 2001, 21, 999–1006. [Google Scholar] [CrossRef]
- Ai, N.; Wood, D.; Yang, E.; Welsh, W.J. Niclosamide is a negative allosteric modulator of group I metabotropic glutamate receptors: Implications for neuropathic pain. Pharmac. Res. 2016, 33, 3044–3056. [Google Scholar] [CrossRef]
- Ocana, M.; Cendan, C.M.; Cobos, E.J.; Entrena, J.M.; Baeyens, J.M. Potassium channels and pain: Present realities and future opportunities. Eur J. Pharm. 2004, 500, 203–219. [Google Scholar] [CrossRef]
- Tsantoulas, C.; McMahon, S.B. Opening paths to novel analgesics: The role of potassium channels in chronic pain. Trends Neurosci. 2014, 37, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Mannion, R.J.; Doubell, T.P.; Coggeshall, R.E.; Woolf, C.J. Collateral sprouting of uninjured primary afferent A-fibers into the superficial dorsal horn of the adult rat spinal cord after topical capsaicin treatment to the sciatic nerve. J. Neurosci. 1996, 16, 5189–5195. [Google Scholar] [CrossRef] [PubMed]
- Mantyh, P.W.; Koltzenburg, M.; Mendell, L.M.; Tive, L.; Shelton, D.L. Antagonism of nerve growth factor-TrkA signaling and the relief of pain. Anesthes 2011, 115, 189–204. [Google Scholar] [CrossRef] [PubMed]
- Nockemann, D.; Rouault, M.; Labuz, D.; Hublitz, P.; McKnelly, K.; Reis, F.C.; Stein, C.; Heppenstall, P.A. The K+ channel GIRK2 is both necessary and sufficient for peripheral opioid-mediated analgesia. EMBO Molec. Med. 2013, 5, 1263–1277. [Google Scholar] [CrossRef]
- Huang, Y.; Zang, Y.; Zhou, L.; Wenshan, G.; Liu, X.; Zhong, Y. The role of TNF-alpha/NF-kappa B pathway on the upregulation of voltage-gated sodium channel Nav1.7 in DRG neurons of rats with diabetic neuropathy. Neurochem. Int. 2014, 75, 112–119. [Google Scholar] [CrossRef]
- Minett, M.S.; Falk, S.; Santana-Varela, S.; Bogdanov, Y.D.; Nassar, M.A.; Heegaard, A.; Wood, J. Pain without nociceptors? Nav1.7-independent pain mechanisms. Cell Rep. 2014, 6, 301–312. [Google Scholar] [CrossRef]
- Xu, H.; Ramsey, I.S.; Kotecha, S.A.; Moran, M.M.; Chong, J.A.; Lawson, D.; Ge, P.; Lilly, J.; Silos-Santiago, I.; Xie, Y.; et al. TRPV3 is a calcium-permeable temperature-sensitive cation channel. Lett. Nat. 2002, 418, 181–186. [Google Scholar] [CrossRef]
- Hu, H.; Xiao, R.; Wang, C.; Gao, N.; Colton, C.K.; Wood, J.D.; Zhu, M.X. Potentiation of TRPV3 channel function by unsaturated fatty acids. J. Cell Physiol. 2006, 208, 201–212. [Google Scholar] [CrossRef] [Green Version]
- Desroches, J.; Bouchard, J.F.; Gendron, P.; Beaulieu, P. Involvement of cannabinoid receptors in peripheral and spinal morphine analgesia. Neurosci 2014, 261, 23–42. [Google Scholar] [CrossRef] [Green Version]
- Rosetti, F.; Mayadas, T.N. The many face of Mac-1 in autoimmune disease. Imunol Rev. 2015, 269, 175–193. [Google Scholar] [CrossRef]
- Moqrich, A.; Hwang, S.W.; Earley, T.J.; Petrus, M.J.; Murray, A.N.; Spencer, K.S.R.; Andahazy, M.; Story, G.M.; Patapoutian, A. Impaired thermosensation in mice lacking TRPV3, a heat and camphor sensor in the skin. Science 2011, 307, 1468–1472. [Google Scholar] [CrossRef] [PubMed]
- Scanzano, A.; Cosentino, M. Adrenergic regulation of innate immunity: A review. Front. Pharm. 2015, 6, 171. [Google Scholar] [CrossRef] [PubMed]
- Abbadle, C.; Lindia, J.A.; Cumiskey, A.M.; Peterson, L.B.; Mudgett, J.S.; Bayne, E.K.; DeMartino, J.A.; MacIntyre, D.E.; Forrest, M.J. Impaired neuropathic pain responses in mice lacking the chemokine receptor CCR2. PNAS 2003, 100, 7947–7952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Zhang, L.; Yu, C.; Yang, X.; Wang, H. Monocyte and macrophage differentiation: Circulation inflammatory monocyte as biomarker for inflammatory disease. Biomark. Res. 2014, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Austin, P.J.; Kim, C.F.; Perera, C.J.; Moalem-Taylor, G. Regulatory T cells attenuate neuropathic pain following peripheral nerve injury and experimental autoimmune neuritis. Pain 2012, 153, 1916–1931. [Google Scholar] [CrossRef] [PubMed]
- Galiegue, S.; Marchand, M.S.; Dussossoy, D.; Carriere, D.; Carayon, P.; Bouaboula, M.; Shire, D.; Le Fur, G.; Casellas, P. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur. J. Biochem. 1995, 232, 54–61. [Google Scholar] [CrossRef]
- Sarafi, M.N.; Garcia-Zepeda, E.A.; MacLean, J.A.; Charo, I.F.; Luster, A.D. Murine monocyte chemoattractant protein (MCP)-5: A novel CC chemokine that is a structural and functional homologue of Human MCP-1. J. Exp. Med. 1997, 185, 99–109. [Google Scholar] [CrossRef]
- Ma, W.; Quirion, R. Does COX2-dependent PGE2 play a role in neuropathic pain? Neurosci. Lett. 2008, 437, 165–169. [Google Scholar] [CrossRef]
- Verge, G.M.; Milligan, E.D.; Maier, S.F.; Watkins, L.R.; Naeve, G.S. Fractalkine (CX3CL1) and fractalkine receptor distribution in spinal cord and dorsal root ganglia under basal and neuropathic conditions. Eur. J. Neurosci. 2004, 20, 1150–1160. [Google Scholar] [CrossRef]
- Hesslinger, C.; Kremmer, E.; Hultner, L.; Ueffing, M.; Irmgard, Z. Phosphorylation of GTP cyclohydrolase I and modulation of its activity in rodent mast cells. J. Biol. Chem. 1998, 273, 21616–21622. [Google Scholar] [CrossRef]
- Bauer, M.; Meyer, M.; Brevig, T.; Gasser, T.; Widmer, H.R.; Zimmer, J.; Ueffing, M. Lipid-mediated glial cell derved neurotrophic factor gene transfer to cultured porcine ventral mesencephalic tissue. Exp. Neurol. 2002, 177, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Lorey, S.L.; Huang, Y.C.; Sharma, V. Constitutive expression of interleukin-18 and interleukin-18 receptor in tumour derived human B-cell lines. Clin. Exp. Immunol. 2004, 136, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Martin, D.P.; Schmelzer, J.D.; Mitsui, Y.; Low, P.A. Pro- and Anti-inflammatory cytokine gene expression in rat sciatic nerve chronic constriction injury model of neuropathic pain. Exper. Neurol. 2001, 169, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; An, J. Cytokines, inflammation and pain. Int. Anesth. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Keyel, P.A. How is inflammation initiated? Individual influences of IL-1, IL-18 and HMGB1. Cytokine 2014, 69, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Hinson, R.M.; Williams, J.A.; Shacter, E. Elevated interleukin 6 is induced by prostaglandin E2 in a murine model of inflammation: Possible role of cyclooxygenase-2. Proc. Natl. Acad. Sci. USA 1996, 93, 4885–4890. [Google Scholar] [CrossRef]
- Xu, Y.; Yoshitake, A.I.; Arai, R. Monoamine oxidase type B is localized to mitochondrial outer membranes in Mast cells, Schwann cells, endothelial cells and fibroblasts of the rat tongue. Acta. Histochem. Cytochem. 2002, 35, 417–422. [Google Scholar] [CrossRef]
- Ma, W.; Eisenbach, J.C. Four PGE2 EP receptors are up-regulated in injured nerve following partial sciatic nerve ligation. Exp. Neurol. 2003, 183, 581–592. [Google Scholar] [CrossRef]
- Lee, S.; Zhang, J. Heterogeneity of macrophages in injured trigeminal nerves: Cytokine/chemokine expressing vs. phagocytic macrophages. Brain Behav. Immun. 2012, 26, 891–903. [Google Scholar] [CrossRef]
- Echeverry, S.; Wu, Y.; Zhang, J. Selectively reducing cytokine/chemokine expressing macrophages in injured nerves impairs the development of neuropathic pain. Exp. Neurol. 2012, 240, 205–218. [Google Scholar] [CrossRef]
- Schafers, M.; Marziniak, M.; Sorkin, L.S.; Yaksh, T.L.; Sommer, C. Cyclooxygenase inhibition in nerve injury- and TNF-induced hyperalgesia in the rat. Exp. Neurol. 2004, 185, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, J.E.; Zygelyte, E.; Grenier, J.K.; Edwards, M.G.; Cheetham, J. Temporal changes in macrophage phenotype after peripheral nerve injury. J. Neuroinflam. 2018, 15, 185. [Google Scholar] [CrossRef] [PubMed]
- Calvo, M.; Dawes, J.M.; Bennett, D.L. The role of the immune system in the generation of neuropathic pain. Lancet. Neurol. 2012, 11, 629–642. [Google Scholar] [CrossRef]
- Ellis, A.; Bennett, D.L.H. Neuroinflammation and the generation of neuropathic pain. Brit. J. Anaes. 2013, 11, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Hirata, T.; Narumiya, S. Prostanoids as regulators of innate and adaptive immunity. Adv. Immunol. 2012, 116, 143–174. [Google Scholar]
- Kalinski, P. Regulation of immune responses by prostaglandin E2. J. Immunol. 2012, 188, 21–28. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, Z.; Chen, S.; Li, M.; Shahveranov, A.; Ye, D.; Tian, Y. Interleukin-6: An emerging regulator of pathological pain. J. Neuroinflam. 2016, 13, 141. [Google Scholar] [CrossRef]
- Hirota, H.; Kiyama, H.; Kishimoto, T.; Taga, T. Accelerated nerve regeneration in mice by upregulated expression of interleukin 6 and IL-6 receptor after trauma. J. Exp. Med. 1996, 183, 2627–2634. [Google Scholar] [CrossRef]
- Makwana, M.; Raivich, G. Molecular mechanisms in successful peripheral regeneration. FEBS 2005, 272, 2628–2638. [Google Scholar] [CrossRef]
- Henken, D.B.; Battisti, W.P.; Chesselet, M.F.; Murray, M.; Tessler, A. Expression of β-preprotachykinin mRNA and tachykinins in rat dorsal root ganglion cells following peripheral or central axotomy. Neuroscience 1990, 39, 733–742. [Google Scholar] [CrossRef]
- Pinto, F.M.; Almeida, T.A.; Hernandez, M.; Devillier, P.; Advenier, C.; Candenas, M.L. mRNA expression of tachykinins and tachykinins receptors in different human tissues. Exper. J. Pharm. 2004, 494, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Ennes, H.S.; McRoberts, J.A.; Marvizon, J.C. Mechanisms of µ-opioid receptor inhibition of NMDA receptor-induced substance P release in the rat spinal cord. Neuropharm 2018, 128, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Kinkelin, I.; Brocker, E.; Koltzenburg, M.; Carlton, S.M. Localization of ionotropic glutamate receptors in peripheral axons of human skin. Neurosci. Lett. 2000, 283, 149–152. [Google Scholar] [CrossRef]
- Sheu, J.Y.; Kulhanek, D.J.; Eckenstein, F.P. Differential patterns of ERK and STAT2 phosphorylation after sciatic nerve transection in the rat. Expl. Neurol. 2000, 166, 392–402. [Google Scholar] [CrossRef]
- Nassar, M.A.; Levato, A.; Stirling, L.C.; Wood, J.N. Neuropathic pain develops normally in mice lacking both Nav1.7 and Nav1.8. Mol. Pain 2005, 1. [Google Scholar] [CrossRef]
- Ferriera 2012 Villarinho, J.G.; Oliveira, S.M.; Silva, C.R.; Cabreira, T.N.; Ferreira, J. Involvement of monoamine oxidase B on models of postoperative and neuropathic pain in mice. Eur. J. Pharm. 2012, 690, 107–114. [Google Scholar] [CrossRef] [Green Version]
- Murali, S.S.; Napier, I.A.; Moahhamdi, S.A.; Alewood, P.F.; Lewis, R.J.; Christie, M.J. High-voltage-activated calcium current subtypes in mouse DRG neurons adapt in a subpopulation-specific manner after nerve injury. J. Neurophysiol. 2015, 113, 1511–1519. [Google Scholar] [CrossRef]
- Staaf, S.; Oerther, S.; Lucas, G.; Mattsson, J.P.; Ernfors, P. Differential regulation of TRP channels in a rat model of neuropathic pain. Pain 2009, 114, 187–199. [Google Scholar] [CrossRef]
- Boucher, T.J.; Okuse, K.; Bennett, D.L.H.; Munson, J.B.; Wood, J.N.; McMahon, S.B. Potent analgesic effects of GDNF in neuropathic pain states. Science 2000, 290, 124–127. [Google Scholar] [CrossRef]
- Takasu, K.; Sakai, A.; Hanawa, H.; Shimada, T.; Suzuki, H. Overexpression of GDNF in the uninjured DRG exerts effects on neuropathic pain following segmental spinal nerve ligation in mice. J. Pain 2011, 12, 1130–1139. [Google Scholar] [CrossRef]
- Yao, J.; Sasaki, Y.; Wen, Z.; Bassell, G.J.; Zheng, J.Q. An essential role for β-actin mRNA localization and translation in Ca2+ -dependent growth cone guidance. Nat. Neurosci. 2006, 9, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shan, Q.; Pan, J.; Yi, S. Actin cytoskeleton affects Schwann cell migration and peripheral nerve regeneration. Front. Physiol. 2018, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Holmes, F.E.; Arnott, N.; Vanderplank, P.; Kerr, N.C.; Longbrake, E.E.; Popovich, P.G.; Imai, T.; Combadiere, C.; Murphy, P.M.; Wynick, D. Intra-neural administration of fractalkine attenuates neuropathic pain-related behavior. J. Neurochem. 2008, 106, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]







| (a). CCI DF-NE vs. Sham. | |||||
|---|---|---|---|---|---|
| Upregulated. CCI DF-NE | Downregulated CCI DF-NE | ||||
| Gene | Fold Regulation | P Value | Gene | Fold Regulation | P Value |
| Actb | 6.61 | 0.0008 | Cacna1b | −6.35 | 0.006 |
| B2m | 1.75 | 0.0002 | Calca | −7.91 | 0.002 |
| Adrb2 | 4.97 | 0.017 | Cckbr | −6.67 | 0.02 |
| Ccl12/Mcp5 | 30.5 | >0.00001 | Grm1 | −6.67 | 0.02 |
| Ccr2 | 2.41 | 0.03 | Grm5 | −6.42 | 0.004 |
| Cd4 | 8.34 | 0.003 | Kcnj6 | −7.16 | 0.02 |
| Cnr2 | 2.68 | 0.045 | Kcnq3 | −5.76 | 0.03 |
| Comt | 2.73 | 0.0004 | Maob | −6.18 | 0.0002 |
| Cx3cr1 | 12.47 | 0.0006 | Mapk3 | −1.62 | 0.016 |
| Ednra1 | 3.59 | 0.018 | Ntrk1 | −7.88 | 0.012 |
| Gch1 | 3.76 | 0.0002 | Oprd1 | −5.03 | 0.005 |
| Gdnf | 1.68 | 0.059 | Oprk1 | −11.03 | 0.048 |
| Il−18 | 6.57 | 0.001 | Opr1m | −10.85 | 0.007 |
| Il−1α | 8.81 | 0.04 | Penk | −5.21 | 0.03 |
| Il−1β | 21.52 | 0.006 | Ptger3 | −2.19 | 0.002 |
| Il−6 | 12.5 | 0.01 | Scn9a | −6.89 | 0.003 |
| Itgam/Cd11b | 12.12 | >0.00001 | |||
| Itgb2/Cd18 | 11.49 | >0.00001 | |||
| P2rx4 | 2.93 | >0.00001 | |||
| P2rx7 | 3.5 | 0.016 | |||
| Prok2 | 5.42 | 0.03 | |||
| Ptger4 | 2.39 | 0.013 | |||
| Tlr2 | 12.22 | >0.00001 | |||
| Tnf | 7.76 | >0.00001 | |||
| Actb | 6.61 | 0.0008 | |||
| B2m | 1.75 | 0.0002 | |||
| (b). CCI CXB-NE vs. Sham. | |||||
| Upregulated CCI CXB-NE | Downregulated CCI CXB-NE | ||||
| Gene | Fold Regulation | P Value | Gene | Fold Regulation | P Value |
| Actb | 8.18 | 0.002 | Cacna1b | −3.03 | 0.02 |
| B2m | 1.84 | 0.009 | Calca | −7.5 | 0.003 0.013 |
| Adrb2 | 4.9 | 0.02 | Edn1 | −2.25 | 0.007 |
| Alox5 | 2.38 | 0.03 | Grm5 | −7.64 | 0.02 |
| Ccl12/Mcp5 | 11.02 | 0.017 | Il−2 | −5.22 | 0.04 |
| Ccr2 | 2.58 | 0.02 | Kcnq3 | −5.7 | 0.00013 |
| Cd4 | 7.35 | 0.01 | Maob | −11.54 | 0.003 |
| Comt | 1.89 | 0.03 | Mapk3 | −2.47 | 0.003 |
| Cx3cr1 | 7.78 | 0.009 | Ntrk1 | −5.98 | 0.03 |
| Gdnf | 4.51 | 0.02 | Oprd1 | −5.31 | 0.01 |
| Il-18 | 4.27 | 0.04 | Oprm1 | −8.02 | 0.01 |
| Il-1β | 6.06 | 0.03 | Ptger3 | −2.46 | 0.005 |
| Il-6 | 3.63 | 0.013 | Scn9a | −14.23 | 0.0009 |
| Itgam/Cd11b | 5.06 | 0.0001 | Trpv3 | −9.12 | 0.0004 |
| Itgb2/Cd18 | 6.84 | 0.02 | |||
| P2rx4 | 2.28 | 0.01 | |||
| Ptger1 | 2.29 | 0.01 | |||
| Tlr2 | 7.12 | 0.01 | |||
| Tnf | 5.64 | 0.02 | |||
| (c). CCI DF-NE vs. CCI CXB-NE. | |||||
| Upregulated CCI DF-NE | Downregulated CCI DF-NE | ||||
| Gene | Fold Regulation | P Value | Gene | Fold Regulation | P Value |
| Grin2b | 2.37 | 0.021 | Cacna1b | −2.1 | 0.04 |
| Il-6 | 3.44 | 0.04 | |||
| Itgam/Cd11b | 2.39 | >0.00001 | |||
| Maob | 1.87 | 0.007 | |||
| Scn9a | 2.06 | 0.05 | |||
| Tac1 | 2.69 | 0.04 | |||
| Trpv3 | 5.33 | 0.01 | |||
| Grin2b | 2.37 | 0.021 | |||
| Gene Symbol | Gene Description | Fold Change Relative to Sham for CCI DF-NE | Fold Change Relative to Sham for CCI CXB-NE | GenBank | Citation |
|---|---|---|---|---|---|
| Actb 1 | Beta-actin | 6.61 | 8.18 | NM_031144 | [41] |
| Cacna1b/Cav2.2 | Calcium channel, voltage-dependent, N type, alpha 1B subunit | 0.16 | 0.33 | NM_147141 | [42] |
| Calca/CGRP | Calcitonin-related polypetide alpha | 0.12 | 0.13 | NM_017338 | [43,44] |
| Comt | Catechol-O-methyltransferase | 2.73 | 1.89 | NM_012531 | [45,46] |
| Ednra | Endothelin receptor type A | 3.59 | 0.44 | NM_012550 | [47] |
| Grm1 | Glutamate receptor, metabotropic 1 | 0.15 | Not detected | NM_017011 | [48] |
| Grm5 | Glutamate receptor, metabotropic 5 | 0.16 | 0.13 | NM_017012 | [48] |
| Kcnj6 | Potassium inwardly-rectifying channel, subfamily J, member 6 | 0.14 | Not detected | NM_013192 | [49,50] |
| Kcnq3 | Potassium voltage-gated channel, KQT-like subfamily, member 3 | 0.17 | 0.18 | NM_133322 | [49,50] |
| Ntrk1/TrkA | Neurotrophic tyrosine kinase receptor, type 1 | 0.13 | 0.17 | NM_021589 | [51,52] |
| Oprd1 | Opioid receptor, delta 1 | 0.2 | 0.19 | NM_012617 | [53] |
| Orpk1 | Opioid receptor, kappa 1 | 0.09 | Not detected | NM_017167 | [53] |
| Oprm1 | Opioid receptor, mu 1 | 0.09 | 0.12 | NM_013071 | [53] |
| P2rx4 | Purinergic receptor P2X, ligand-gated ion channel 4 | 2.96 | 2.28 | NM_031594 | [54] |
| Scn9a/Nav1.7 | Sodium channel, voltage-gated, type IX, alpha | 0.15 | 0.07 | NM_133289 | [55] |
| TrpV3 | Transient receptor potential cation channel, subfamily V, member 3 | Not detected | 0.11 | NM_001025757 | [56,57,58] |
| Gene Symbol | Gene Description | Fold Change Relative to Sham for CCI DF-NE | Fold Change Relative to Sham for CCI CXB-NE | Cell Expression | GenBank | Citation |
|---|---|---|---|---|---|---|
| Adrb2 | Adrenergic beta-2 receptor | 4.97 | 4.9 | Macrophages, Circulating monocytes | NM_012492 | [61] |
| Ccr2 | Chemokine (C-C) receptor 2 | 2.41 | 2.58 | Macrophages | NM_021866 | [62,63] |
| Cd4 1 | CD4 cell | 8.34 | 7.35 | T Cells | NM_012705 | [64] |
| Cnr2 | Cannabinoid receptor 2 | 2.68 | Not detected | Macrophages | NM_012784 | [65,66] |
| Itgam/Cd11b 1 | Integrin, alpha M | 12.12 | 5.06 | Macrophages, Circulating monocytes | NM_012711 | [59] |
| Itgb2/Cd18 1 | Integrin, beta 2 | 11.49 | 6.84 | Macrophages, Circulating monocytes | NM_001037780 | [59] |
| Mcp5/Ccl12 1 | Chemokine (C-C motif) ligand 12 | 30.5 | 11.02 | Macrophages | NM_001105822 | [60] |
| Ptgs2 | Prostaglandin E synthase 2 | Not detected | 2.21 | Macrophages, Circulating monocytes | NM_001107832 | [67] |
| Gene Symbol | Gene Description | Fold Change Relative to Sham for CCI DF-NE | Fold Change Relative to Sham for CCI CXB-NE | Cell Expression | GenBank | Citation |
|---|---|---|---|---|---|---|
| Cx3cr1 1 | Chemokine (C-X3-C motif) receptor 1 | 12.47 | 7.78 | Macrophages, circulating monocytes, neuronal, T cells | NM_133534 | [16,68] |
| Gch1 | GTP cyclohydrolase 1 | 3.76 | Not detected | Macrophages, neuronal, T cells, mast cells | NM_024356 | [69] |
| Gdnf 1 | Glial cell derived neurotrophic factor | 1.68 | 4.51 | Neuronal, Schwann cells | NM_019139 | [70] |
| IL-18 1 | Interleukin 18 | 6.57 | 4.27 | Macrophages, circulating monocytes, neuronal, Schwann cells | NM_019165 | [38,71] |
| IL-1β 1 | Interleukin 1 beta | 21.52 | 6.06 | Macrophages, circulating monocytes, neuronal, endothelial cells | NM_031512 | [72,73,74] |
| IL-6 1 | Interleukin 6 | 12.5 | 3.63 | Macrophages, circulating monocytes, Schwann cells | NM_012589 | [72,75] |
| Maob | Monoamine oxidase B | 0.16 | 0.09 | Neuronal, Schwann cells, endothelial cells | NM_013198 | [76] |
| Mapk3 | Mitogen-activated protein kinase 3 | 0.62 | 0.40 | Neuronal, Schwann cells | NM_017347 | [39] |
| Ptger1 | Prostaglandin E receptor 1 | Not detected | 2.29 | Macrophages, neuronal | NM_013100 | [77] |
| Ptger3 | Prostaglandin E receptor 3 | 0.46 | 0.41 | Macrophages, neuronal | NM_012704 | [77] |
| Ptger4 | Prostaglandin E receptor 4 | 2.39 | 2.14 | Macrophages, neuronal | NM_032076 | [77] |
| Tlr2 | Toll-like receptor 2 | 12.22 | 7.12 | Macrophages, circulating monocytes, Schwann cells, T cells | NM__198769 | [13] |
| Tnf | Tumor necrosis factor | 7.76 | 5.64 | Macrophages, circulating monocytes, Schwann cells, T cells | NM_012675 | [72] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stevens, A.M.; Liu, L.; Bertovich, D.; Janjic, J.M.; Pollock, J.A. Differential Expression of Neuroinflammatory mRNAs in the Rat Sciatic Nerve Following Chronic Constriction Injury and Pain-Relieving Nanoemulsion NSAID Delivery to Infiltrating Macrophages. Int. J. Mol. Sci. 2019, 20, 5269. https://doi.org/10.3390/ijms20215269
Stevens AM, Liu L, Bertovich D, Janjic JM, Pollock JA. Differential Expression of Neuroinflammatory mRNAs in the Rat Sciatic Nerve Following Chronic Constriction Injury and Pain-Relieving Nanoemulsion NSAID Delivery to Infiltrating Macrophages. International Journal of Molecular Sciences. 2019; 20(21):5269. https://doi.org/10.3390/ijms20215269
Chicago/Turabian StyleStevens, Andrea M., Lu Liu, Dylan Bertovich, Jelena M. Janjic, and John A. Pollock. 2019. "Differential Expression of Neuroinflammatory mRNAs in the Rat Sciatic Nerve Following Chronic Constriction Injury and Pain-Relieving Nanoemulsion NSAID Delivery to Infiltrating Macrophages" International Journal of Molecular Sciences 20, no. 21: 5269. https://doi.org/10.3390/ijms20215269
APA StyleStevens, A. M., Liu, L., Bertovich, D., Janjic, J. M., & Pollock, J. A. (2019). Differential Expression of Neuroinflammatory mRNAs in the Rat Sciatic Nerve Following Chronic Constriction Injury and Pain-Relieving Nanoemulsion NSAID Delivery to Infiltrating Macrophages. International Journal of Molecular Sciences, 20(21), 5269. https://doi.org/10.3390/ijms20215269