Discovery of New Cyclopentaquinoline Analogues as Multifunctional Agents for the Treatment of Alzheimer’s Disease
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biological Evaluation
2.2.1. In Vitro Inhibition Studies on AChE and BuChE
2.2.2. Kinetic Evaluation of Compound 3e
2.2.3. β-Amyloid Assay
2.2.4. In Vitro Cytotoxicity Assay
2.2.5. In Vitro Hepatotoxicity
2.2.6. In Vitro Inhibition Study on Hyaluronidase (HYAL)
2.2.7. LogP and pKa Assay
2.2.8. Yeast Three-Hybrid Technology (Y3H) Test
2.2.9. ADMET (Absorption, Distribution, Metabolism, Excretion, Toxicity) Analysis
2.2.10. Antioxidant Activity Assay
2.2.11. Neuroprotection Against Oxidative Stress
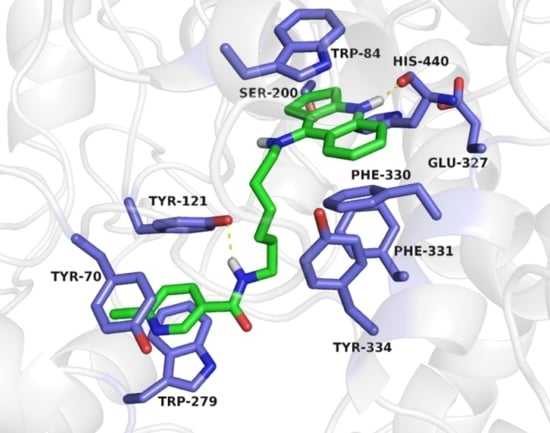
2.2.12. Molecular Modeling
3. Materials and Methods
3.1. Synthesis
3.1.1. General Procedure for the Synthesis of Compound 1a–1h
3.1.2. General Procedure for the Synthesis of Compounds 2a–2h
6-chloro-N-[2-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)ethyl]pyridine-3-carboxamide (2a)
6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)propyl]pyridine-3-carboxamide (2b)
6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)butyl]pyridine-3-carboxamide (2c)
6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)pentyl]pyridine-3-carboxamide (2d)
6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)hexyl]pyridine-3-carboxamide (2e)
6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)heptyl]pyridine-3-carboxamide (2f)
6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)octyl]pyridine-3-carboxamide (2g)
6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)nonyl]pyridine-3-carboxamide (2h)
3.2. General Procedure for the Synthesis of Compounds 3a–3h
3.2.1. 6-chloro-N-[2-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)ethyl]pyridine-3-carboxamide hydrochloride (3a)
3.2.2. 6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)propyl]pyridine-3-carboxamide hydrochloride (3b)
3.2.3. 6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)butyl]pyridine-3-carboxamide hydrochloride (3c)
3.2.4. 6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)pentyl]pyridine-3-carboxamide hydrochloride (3d)
3.2.5. 6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)hexyl]pyridine-3-carboxamide hydrochloride (3e)
3.2.6. 6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)heptyl]pyridine-3-carboxamide hydrochloride (3f)
3.2.7. 6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)octyl]pyridine-3-carboxamide hydrochloride (3g)
3.2.8. 6-chloro-N-[3-(2,3-dihydro-1H-cyclopenta[b]quinolin-9-ylamino)nonyl]pyridine-3-carboxamide hydrochloride (3h)
3.3. Biochemical Evaluation
3.3.1. In Vitro Inhibition Studies on AChE and BuChE
3.3.2. Kinetic Characterization of AChE Inhibition
3.3.3. Beta-Amyloid Assay
3.3.4. Cell Culture and Cytotoxicity Assay
3.3.5. Cell Culture and Determination of Hepatotoxicity
3.3.6. Hyaluronidase Inhibition Test
3.3.7. pKa Assay
3.3.8. LogP Assay
3.3.9. A Yeast Three-Hybrid (Y3H) Technology Test
3.3.10. ADMET Analysis
3.3.11. Antioxidant Activity Assay
3.3.12. Neuroprotection Against Oxidative Stress
Cell Culture
MTT Assay
3.3.13. Molecular Modeling
4. Conclusions
Author Contributions
Acknowledgment
Conflicts of Interest
References
- Goedert, M.; Spillantini, M.G. A century of Alzheimer’s disease. Science 2006, 314, 777–781. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.; He, L.; Mak, M.; Han, Y.; Ho, C.-Y.; Zuo, Z. Synthesis, biological activity, and biopharmaceutical characterization of tacrine dimers as acetylcholinesterase inhibitors. Int. J. Pharm. 2014, 477, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, E.; Chin, T.Y.; Chen, C.P.; Wu, T.Y. Management of moderate to severe Alzheimer’s disease: Focus on memantine. Taiwan J. Obstet. Gynecol. 2011, 50, 415–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evin, G.; Barakat, A.; Masters, C.L. Bace: Therapeutic target and potential biomarker for Alzheimer’s disease. Int. J. Biochem. Cell Biol. 2010, 42, 1923–1926. [Google Scholar] [CrossRef] [PubMed]
- Terry, A.V., Jr.; Buccafusco, J.J. The cholinergic hypothesis of age and Alzheimer’s disease-related cognitive deficits: Recent challenges and their implications for novel drug development. J. Pharmacol. Exp. Ther. 2003, 306, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, A.; Bolognesi, M.L.; Minarini, A.; Rosini, M.; Tumiatti, V.; Recanatini, M.; Melchiorre, C. Multi-target-directed ligands to combat neurodegenerative diseases. J. Med. Chem. 2008, 51, 347–372. [Google Scholar] [CrossRef] [PubMed]
- Wieckowska, A.; Wieckowski, K.; Bajda, M.; Brus, B.; Salat, K.; Czerwinska, P.; Gobec, S.; Filipek, B.; Malawska, B. Synthesis of new N-benzylpiperidine derivatives as cholinesterase inhibitors with beta-amyloid anti-aggregation properties and beneficial effects on memory in vivo. Bioorg. Med. Chem. 2015, 23, 2445–2457. [Google Scholar] [CrossRef]
- Poprac, P.; Jomova, K.; Simunkova, M.; Kollar, V.; Rhodes, C.J.; Valko, M. Targeting free radicals in oxidative stress-related human diseases. Trends Pharmacol. Sci. 2017, 38, 592–607. [Google Scholar] [CrossRef]
- Davies, A.M.; Holt, A.G. Why antioxidant therapies have failed in clinical trials. J. Theor. Biol. 2018, 457, 1–5. [Google Scholar] [CrossRef]
- Szymański, P.; Lázničková, A.; Lázniček, M.; Bajda, M.; Malawska, B.; Markowicz, M.; Mikiciuk-Olasik, E. 2,3-dihydro-1h-cyclopenta[b]quinoline derivatives as acetylcholinesterase inhibitors-synthesis, radiolabeling and biodistribution. Int. J. Mol. Sci. 2012, 13, 10067–10090. [Google Scholar] [CrossRef]
- Czarnecka, K.; Szymański, P.; Girek, M.; Mikiciuk-Olasik, E.; Skibiński, R.; Kabziński, J.; Majsterek, I.; Malawska, B.; Jończyk, J.; Bajda, M. Tetrahydroacridine derivatives with fluorobenzoic acid moiety as multifunctional agents for Alzheimer’s disease treatment. Bioorg. Chem. 2017, 72 (Suppl. C), 315–322. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L.; Courtney, K.D.; Andres, V.; Featherstone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef] [Green Version]
- Czarnecka, K.; Girek, M.; Maciejewska, K.; Skibiński, R.; Jończyk, J.; Bajda, M.; Kabziński, J.; Sołowiej, P.; Majsterek, I.; Szymański, P. New cyclopentaquinoline hybrids with multifunctional capacities for the treatment of Alzheimer’s disease. J. Enzym. Inhib. Med. Chem. 2017, 33, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.T.; Almeida, J.R.; Araujo, A.A.; Duarte, M.C.; Gelain, D.P.; Moreira, J.C.; dos Santos, M.R.; Quintans-Junior, L.J. Structure-activity relationship of terpenes with anti-inflammatory profile—A systematic review. Basic Clin. Pharmacol. Toxicol. 2014, 115, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Salvamani, S.; Gunasekaran, B.; Shukor, M.Y.; Shaharuddin, N.A.; Sabullah, M.K.; Ahmad, S.A. Anti-hmg-coa reductase, antioxidant, and anti-inflammatory activities of amaranthus viridis leaf extract as a potential treatment for hypercholesterolemia. Evid. Based Complement. Alternat. Med. 2016, 2016, 10. [Google Scholar] [CrossRef] [PubMed]
- Hansch, C.; Leo, A.; Hoekman, D. Exploring Qsar: Hydrophobic, Electronic, and Steric Constants; American Chemical Society: Washington, DC, USA, 1995. [Google Scholar]
- Sangster, J. Octanol-Water Partition Coefficients: Fundamentals and Physical Chemistry; Wiley: Chichester, UK, 1997. [Google Scholar]
- Kortum, G.; Vogel, W.; Andrussow, K. International Union of Pure and Applied Chemistry; Commission on Electrochemical Data. In Dissociation Constants of Organic Acids in Aqueous Solution; Butterworths: London, UK, 1961. [Google Scholar]
- Perrin, D.D. Dissociation Constants of Organic Bases in Aqueous Solution; IUPAC Chemical Data Series: Supplement 1972; Butterworths: London, UK, 1972. [Google Scholar]
- O’neil, M.J.; Heckelman, P.E.; Koch, C.B.; Roman, K.J. (Eds.) The Merck Index: An Encyclopedia of Chemicals, Drugs, and Biologicals, 14th ed.; Merck and Co., Inc.: Whitehouse Station, NJ, USA, 2006; 2564p, ISBN 0-911910-00-x. [Google Scholar]
- Newhouse, K.; Hsuan, S.L.; Chang, S.H.; Cai, B.; Wang, Y.; Xia, Z. Rotenone-induced apoptosis is mediated by p38 and JNK MAP kinases in human dopaminergic SH-SY5Y cells. Toxicol. Sci. 2004, 79, 137–146. [Google Scholar] [CrossRef]
- Romero, A.; Egea, J.; Gonzalez-Munoz, G.C.; Martin de Saavedra, M.D.; del Barrio, L.; Rodriguez-Franco, M.I.; Conde, S.; Lopez, M.G.; Villarroya, M.; de los Rios, C. ITH12410/SC058: A new neuroprotective compound with potential in the treatment of Alzheimer’s disease. ACS Chem. Neurosci. 2014, 5, 770–775. [Google Scholar] [CrossRef]
- González-Muñoz, G.C.; Arce, M.P.; López, B.; Pérez, C.; Romero, A.; Barrio, L.D.; Martín-de-Saavedra, M.D.; Egea, J.; León, R.; Villarroya, M.; et al. N-acylaminophenothiazines: Neuroprotective agents displaying multifunctional activities for a potential treatment of Alzheimer’s disease. Eur. J. Med. Chem. 2011, 46, 2224–2235. [Google Scholar] [CrossRef]
- Bartolini, M.; Bertucci, C.; Bolognesi, M.L.; Cavalli, A.; Melchiorre, C.; Andrisano, V. Insight into the kinetic of amyloid beta (1-42) peptide self-aggregation: Elucidation of inhibitors’ mechanism of action. Chembiochem 2007, 8, 2152–2161. [Google Scholar] [CrossRef]
- Keri, R.S.; Quintanova, C.; Chaves, S.; Silva, D.F.; Cardoso, S.M.; Santos, M.A. New tacrine hybrids with natural-based cysteine derivatives as multitargeted drugs for potential treatment of Alzheimer’s disease. Chem. Biol. Drug Des. 2016, 87, 101–111. [Google Scholar] [CrossRef]
- Camps, P.; Formosa, X.; Galdeano, C.; Munoz-Torrero, D.; Ramirez, L.; Gomez, E.; Isambert, N.; Lavilla, R.; Badia, A.; Clos, M.V.; et al. Pyrano[3,2-c]quinoline-6-chlorotacrine hybrids as a novel family of acetylcholinesterase- and beta-amyloid-directed anti-Alzheimer compounds. J. Med. Chem. 2009, 52, 5365–5379. [Google Scholar] [CrossRef] [PubMed]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di Virgilio, A.; Conticello, C.; Ruco, L.; Peschle, C.; De Maria, R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2007, 15, 504–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majsterek, I.; Blasiak, J.; Mlynarski, W.; Hoser, G.; Skorski, T. Does the bcr/abl-mediated increase in the efficacy of DNA repair play a role in the drug resistance of cancer cells? Cell Biol. Int. 2002, 26, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.V.; Tan, A.S. Characterization of the cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT): Subcellular localization, substrate dependence, and involvement of mitochondrial electron transport in MTT reduction. Arch. Biochem. Biophys. 1993, 303, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Plumb, J.A. Cell sensitivity assays: The MTT assay. In Cancer Cell Culture: Methods and Protocols; Langdon, S.P., Ed.; Humana Press: Totowa, NJ, USA, 2004; pp. 165–169. [Google Scholar]
- Mao, F.; Li, J.; Wei, H.; Huang, L.; Li, X. Tacrine-propargylamine derivatives with improved acetylcholinesterase inhibitory activity and lower hepatotoxicity as a potential lead compound for the treatment of Alzheimer’s disease. J. Enzym. Inhib. Med. Chem. 2015, 30, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zenger, K.; Lupp, A.; Kling, B.; Heilmann, J.; Fleck, C.; Kraus, B.; Decker, M. Tacrine-silibinin codrug shows neuro- and hepatoprotective effects in vitro and pro-cognitive and hepatoprotective effects in vivo. J. Med. Chem. 2012, 55, 5231–5242. [Google Scholar] [CrossRef] [PubMed]
- Michel, P.; Owczarek, A.; Matczak, M.; Kosno, M.; Szymański, P.; Mikiciuk-Olasik, E.; Kilanowicz, A.; Wesołowski, W.; Olszewska, A.M. Metabolite profiling of eastern teaberry (Gaultheria procumbens L.) lipophilic leaf extracts with hyaluronidase and lipoxygenase inhibitory activity. Molecules 2017, 22, 412. [Google Scholar] [CrossRef] [PubMed]
- Musil, K.; Florianova, V.; Bucek, P.; Dohnal, V.; Kuca, K.; Musilek, K. Development and validation of a FIA/UV–vis method for pKa determination of oxime based acetylcholinesterase reactivators. J. Pharm. Biomed. Anal. 2016, 117, 240–246. [Google Scholar] [CrossRef]
- Liang, C.; Qiao, J.Q.; Lian, H.Z. Determination of reversed-phase high performance liquid chromatography based octanol-water partition coefficients for neutral and ionizable compounds: Methodology evaluation. J. Chromatogr. A 2017, 1528, 25–34. [Google Scholar] [CrossRef]
- Blois, M.S. Antioxidant determinations by the use of a stable free radical. Nature 1958, 181, 1199. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved abts radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Corina. Sybyl 8.0 (Tripos). Available online: http://www.molecular-networks.com/online_demos/corina_demo (accessed on 1 June 2018).
- Sybyl-x 1.1; Tripos: St. Louis, MO, USA, 2010.
- Gold 5.1; The Cambridge Crystallographic Data Centre: Cambridge, UK, 2011.
- Pymol 0.99rc6; Delano Scientific LLC: Palo Alto, CA, USA, 2006.
- Causier, B.; Davies, B. Analysing protein-protein interactions with the yeast two-hybrid system. Plant Mol. Biol. 2002, 50, 855–870. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, A.; Colas, P. Yeast two-hybrid methods and their applications in drug discovery. Trends Pharmacol. Sci. 2012, 33, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16s primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Cottier, S.; Monig, T.; Wang, Z.; Svoboda, J.; Boland, W.; Kaiser, M.; Kombrink, E. The yeast three-hybrid system as an experimental platform to identify proteins interacting with small signaling molecules in plant cells: Potential and limitations. Front. Plant Sci. 2011, 2, 101. [Google Scholar] [CrossRef] [PubMed]
- Henthorn, D.C.; Jaxa-Chamiec, A.A.; Meldrum, E. A GAL4-based yeast three-hybrid system for the identification of small molecule-target protein interactions. Biochem. Pharmacol. 2002, 63, 1619–1628. [Google Scholar] [CrossRef]
- Licitra, E.J.; Liu, J.O. A three-hybrid system for detecting small ligand-protein receptor interactions. Proc. Natl. Acad. Sci. USA 1996, 93, 12817–12821. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Comp. | AChE IC50 ± SD (µM) a | BuChE IC50 ± SD (µM) b | Selectivity for AChE c | Selectivity for BuChE d | ABTS FRS50 ± SD (mM) | DPPH FRS50 ± SD (mM) |
|---|---|---|---|---|---|---|
| 3a | 0.453 ± 0.018 | 0.124 ± 0.037 | 0.3 | 3.6 | 3.1 ± 0.18 | 14.3 ± 1.46 |
| 3b | 0.245 ± 0.038 | 0.618 ± 0.043 | 2.5 | 0.4 | 1.9 ± 0.30 | NA |
| 3c | 0.589 ± 0.072 | 0.662 ± 0.053 | 1.1 | 0.9 | 3.6 ± 0.66 | NA |
| 3d | 0.162 ± 0.018 | 0.534 ± 0.023 | 3.3 | 0.3 | 4.8 ± 0.66 | NA |
| 3e | 0.067 ± 0.002 | 0.153 ± 0.020 | 2.3 | 0.4 | NA | NA |
| 3f | 0.074 ± 0.004 | 0.062 ± 0.003 | 0.8 | 1.2 | 10.5 ± 1.79 | NA |
| 3g | 0.073 ± 0.005 | 0.042 ± 0.006 | 0.6 | 1.2 | 4.8 ± 0.34 | NA |
| 3h | 0.071 ± 0.005 | 0.094 ± 0.006 | 1.3 | 0.8 | NA | NA |
| THA | 0.081 ± 0.009 | 0.020 ± 0.002 | 0.24 | 4.13 | NA | NA |
| Trolox | - | - | - | - | 0.040 ± 0.006 | 0.0049 ± 0.0004 |
| 3e Compound Concentration (µM) | Inhibition of Aβ Aggregation (%) |
|---|---|
| Control sample | 0.00 |
| 10 | 40 ± 4.8 |
| 25 | 63 ± 4.2 |
| 50 | 72 ± 2.9 |
| 100 | 79 ± 3.2 |
| Parameter | Experimental | ChemAxon | ACD/Percepta |
|---|---|---|---|
| pKa1 | 8.53 | 8.89 | 9.01 |
| pKa2 | 11.34 | 13.78 | 13.74 |
| logP | 3.990 | 4.43 | 5.00 |
| Substance Name | Experimental Properties | |
|---|---|---|
| logP [16] | pKa | |
| Procainamide HCl | 0.88 | 9.32 [17] |
| Salicynamide | 1.28 | 8.37 [18] |
| Tacrine | 2.71 | 9.95 [19] |
| Thymol | 3.3 | - |
| Naphtalene | 3.6 | - |
| Promazine HCl | 4.55 | 9.36 [17] |
| Prometazine HCl | 4.81 | 9.1 [17] |
| Chlorpromazine HCl | 5.41 | 9.3 [16] |
| Thiorydazine HCl | 5.9 | 9.5 [20] |
| Comp. | Concentration | 10 µM | 1 µM | 0.1 µM | 0.01 µM | |
|---|---|---|---|---|---|---|
| H2O2 100 µM | % cell viability | |||||
| 3e | 14.3 ± 1.42 | 18.4 ± 2.15 | 29.2 ± 0.41 | 60.4 ± 3.13 | ||
| Trolox | 99.2 ± 3.30 | 99.4 ± 9.81 | 98.6 ± 1.36 | 96.4 ± 1.51 | ||
| Comp. | Concentration | 0.1 µM | 0.01 µM | 0.001 µM | 0.0001 µM | |
| Rotenone/Oligomycin A 30/10 µM | % cell viability | |||||
| 3e | Pre-incubation | 34.2 ± 1.73 | 40.2 ± 2.13 | 44.5 ± 3.52 | 48.9 ± 4.42 | |
| Co-incubation | 40.6 ± 4.91 | 42.6 ± 4.87 | 43.9 ± 4.76 | 45.6 ± 3.51 | ||
| Trolox | Pre-incubation | 44.3 ± 3.73 | 46.4 ± 2.06 | 47.9 ± 4.48 | 46.2 ± 3.65 | |
| Co-incubation | 48.1 ± 8.01 | 50.3 ± 8.11 | 50.8 ± 6.98 | 42.1 ± 6.28 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czarnecka, K.; Girek, M.; Kręcisz, P.; Skibiński, R.; Łątka, K.; Jończyk, J.; Bajda, M.; Kabziński, J.; Majsterek, I.; Szymczyk, P.; et al. Discovery of New Cyclopentaquinoline Analogues as Multifunctional Agents for the Treatment of Alzheimer’s Disease. Int. J. Mol. Sci. 2019, 20, 498. https://doi.org/10.3390/ijms20030498
Czarnecka K, Girek M, Kręcisz P, Skibiński R, Łątka K, Jończyk J, Bajda M, Kabziński J, Majsterek I, Szymczyk P, et al. Discovery of New Cyclopentaquinoline Analogues as Multifunctional Agents for the Treatment of Alzheimer’s Disease. International Journal of Molecular Sciences. 2019; 20(3):498. https://doi.org/10.3390/ijms20030498
Chicago/Turabian StyleCzarnecka, Kamila, Małgorzata Girek, Paweł Kręcisz, Robert Skibiński, Kamil Łątka, Jakub Jończyk, Marek Bajda, Jacek Kabziński, Ireneusz Majsterek, Piotr Szymczyk, and et al. 2019. "Discovery of New Cyclopentaquinoline Analogues as Multifunctional Agents for the Treatment of Alzheimer’s Disease" International Journal of Molecular Sciences 20, no. 3: 498. https://doi.org/10.3390/ijms20030498
APA StyleCzarnecka, K., Girek, M., Kręcisz, P., Skibiński, R., Łątka, K., Jończyk, J., Bajda, M., Kabziński, J., Majsterek, I., Szymczyk, P., & Szymański, P. (2019). Discovery of New Cyclopentaquinoline Analogues as Multifunctional Agents for the Treatment of Alzheimer’s Disease. International Journal of Molecular Sciences, 20(3), 498. https://doi.org/10.3390/ijms20030498