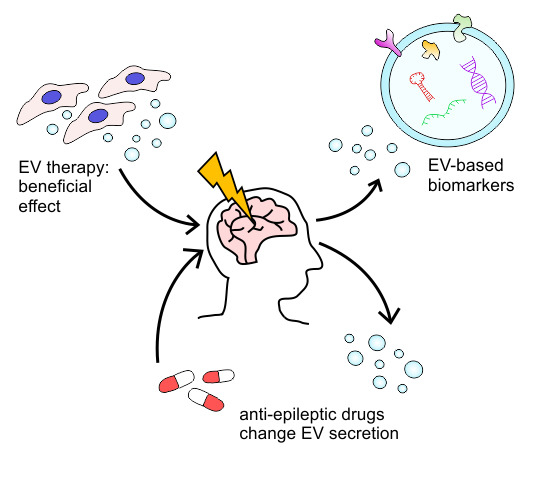
Extracellular Vesicles as Diagnostics and Therapeutics for Structural Epilepsies
Abstract
:1. Introduction
2. Methodological Challenges in EV Isolation and Cargo Analysis in Liquid Biopsies
3. Heterogeneity of EVs
4. Extracellular Vesicle Profile in Plasma and CSF during the Epileptogenic Process
5. EV number in Plasma and CSF during Epileptogenesis after TBI
5.1. Experimental TBI
5.1.1. CCI
5.1.2. Lateral FPI
5.1.3. Other Models
5.2. Human TBI
5.2.1. Plasma
5.2.2. CSF
5.3. Caveats Related to Analysis EVs after TBI
6. Regulation of Body-Fluid EV Cargo during Epileptogenic Process with Focus on microRNAs
6.1. Post-Traumatic Epileptogenesis
6.2. Epilepsy
7. EV-Related Transcripts are Positively Enriched in Rodent Models of Epilepsy and Epileptogenesis
7.1. Post-Traumatic Epileptogenesis
7.2. Epilepsy
8. EV Therapy Improves the Functional Outcome after Status Epilepticus and TBI
8.1. Post-Traumatic Epileptogenesis
8.2. Epilepsy
9. Anti-Seizure Drugs Modulate the Expression of Genes Related to EVs
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Stafstrom, C.E.; Carmant, L. Seizures and epilepsy: An overview for neuroscientists. Cold Spring Harb. Perspect. Med. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Pitkanen, A.; Engel, J., Jr. Past and present definitions of epileptogenesis and its biomarkers. Neurotherapeutics 2014, 11, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Pitkanen, A.; Ekolle Ndode-Ekane, X.; Lapinlampi, N.; Puhakka, N. Epilepsy biomarkers—Toward etiology and pathology specificity. Neurobiol. Dis. 2018, 123, 42–58. [Google Scholar] [CrossRef] [PubMed]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, J.; Wysoczynski, M.; Hayek, F.; Janowska-Wieczorek, A.; Ratajczak, M.Z. Membrane-derived microvesicles: Important and underappreciated mediators of cell-to-cell communication. Leukemia 2006, 20, 1487–1495. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subra, C.; Grand, D.; Laulagnier, K.; Stella, A.; Lambeau, G.; Paillasse, M.; De Medina, P.; Monsarrat, B.; Perret, B.; Silvente-Poirot, S.; et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 2010, 51, 2105–2120. [Google Scholar] [CrossRef] [Green Version]
- Guescini, M.; Genedani, S.; Stocchi, V.; Agnati, L.F. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J. Neural Transm. 2010, 117, 1–4. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [Green Version]
- Laulagnier, K.; Motta, C.; Hamdi, S.; Roy, S.; Fauvelle, F.; Pageaux, J.F.; Kobayashi, T.; Salles, J.P.; Perret, B.; Bonnerot, C.; et al. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 2004, 380, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. The potential of tumor-derived exosomes for noninvasive cancer monitoring. Expert Rev. Mol. Diagn. 2015, 15, 1293–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jan, A.T.; Malik, M.A.; Rahman, S.; Yeo, H.R.; Lee, E.J.; Abdullah, T.S.; Choi, I. Perspective Insights of Exosomes in Neurodegenerative Diseases: A Critical Appraisal. Front Aging Neurosci. 2017, 9, 317. [Google Scholar] [CrossRef] [PubMed]
- Kanninen, K.M.; Bister, N.; Koistinaho, J.; Malm, T. Exosomes as new diagnostic tools in CNS diseases. Biochim. Biophys. Acta 2016, 1862, 403–410. [Google Scholar] [CrossRef]
- Armstrong, D.; Wildman, D.E. Extracellular Vesicles and the Promise of Continuous Liquid Biopsies. J. Pathol. Transl. Med. 2018, 52, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Giebel, B.; Kordelas, L.; Borger, V. Clinical potential of mesenchymal stem/stromal cell-derived extracellular vesicles. Stem Cell Investig. 2017, 4, 84. [Google Scholar] [CrossRef]
- Lai, R.C.; Arslan, F.; Lee, M.M.; Sze, N.S.; Choo, A.; Chen, T.S.; Salto-Tellez, M.; Timmers, L.; Lee, C.N.; El Oakley, R.M.; et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010, 4, 214–222. [Google Scholar] [CrossRef] [Green Version]
- Webb, R.L.; Kaiser, E.E.; Scoville, S.L.; Thompson, T.A.; Fatima, S.; Pandya, C.; Sriram, K.; Swetenburg, R.L.; Vaibhav, K.; Arbab, A.S.; et al. Human Neural Stem Cell Extracellular Vesicles Improve Tissue and Functional Recovery in the Murine Thromboembolic Stroke Model. Transl. Stroke Res. 2018, 9, 530–539. [Google Scholar] [CrossRef]
- El-Andaloussi, S.; Lee, Y.; Lakhal-Littleton, S.; Li, J.; Seow, Y.; Gardiner, C.; Alvarez-Erviti, L.; Sargent, I.L.; Wood, M.J. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat. Protoc. 2012, 7, 2112–2126. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Kramer-Albers, E.M. Ticket to Ride: Targeting Proteins to Exosomes for Brain Delivery. Mol. Ther. 2017, 25, 1264–1266. [Google Scholar] [CrossRef]
- Van der Pol, E.; Boing, A.N.; Gool, E.L.; Nieuwland, R. Recent developments in the nomenclature, presence, isolation, detection and clinical impact of extracellular vesicles. J. Thromb. Haemost. 2016, 14, 48–56. [Google Scholar] [CrossRef]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013, 113, 1–11. [Google Scholar] [CrossRef]
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Witwer, K.W.; Soekmadji, C.; Hill, A.F.; Wauben, M.H.; Buzas, E.I.; Di Vizio, D.; Falcon-Perez, J.M.; Gardiner, C.; Hochberg, F.; Kurochkin, I.V.; et al. Updating the MISEV minimal requirements for extracellular vesicle studies: Building bridges to reproducibility. J. Extracell. Vesicles 2017, 6, 1396823. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- EV-TRACK Consortium; Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, O.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; Bertier, L.; et al. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 2017, 14, 228–232. [Google Scholar] [CrossRef]
- Gardiner, C.; Di Vizio, D.; Sahoo, S.; Thery, C.; Witwer, K.W.; Wauben, M.; Hill, A.F. Techniques used for the isolation and characterization of extracellular vesicles: Results of a worldwide survey. J. Extracell. Vesicles 2016, 5, 32945. [Google Scholar] [CrossRef]
- Simonsen, J.B. What Are We Looking At? Extracellular Vesicles, Lipoproteins, or Both? Circ. Res. 2017, 121, 920–922. [Google Scholar] [CrossRef]
- Karimi, N.; Cvjetkovic, A.; Jang, S.C.; Crescitelli, R.; Hosseinpour Feizi, M.A.; Nieuwland, R.; Lotvall, J.; Lasser, C. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell. Mol. Life Sci. 2018, 75, 2873–2886. [Google Scholar] [CrossRef] [Green Version]
- Sodar, B.W.; Kittel, A.; Paloczi, K.; Vukman, K.V.; Osteikoetxea, X.; Szabo-Taylor, K.; Nemeth, A.; Sperlagh, B.; Baranyai, T.; Giricz, Z.; et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci. Rep. 2016, 6, 24316. [Google Scholar] [CrossRef] [Green Version]
- Karttunen, J.; Heiskanen, M.; Navarro-Ferrandis, V.; Das Gupta, S.; Lipponen, A.; Puhakka, N.; Rilla, K.; Koistinen, A.; Pitkanen, A. Precipitation-based extracellular vesicle isolation from rat plasma co-precipitate vesicle-free microRNAs. J. Extracell. Vesicles 2018, 8, 1555410. [Google Scholar] [CrossRef]
- Yuana, Y.; Levels, J.; Grootemaat, A.; Sturk, A.; Nieuwland, R. Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef]
- Gardiner, C.; Ferreira, Y.J.; Dragovic, R.A.; Redman, C.W.; Sargent, I.L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Mork, M.; Pedersen, S.; Botha, J.; Lund, S.M.; Kristensen, S.R. Preanalytical, analytical, and biological variation of blood plasma submicron particle levels measured with nanoparticle tracking analysis and tunable resistive pulse sensing. Scand. J. Clin. Lab Investig. 2016, 76, 349–360. [Google Scholar] [CrossRef]
- Perez-Gonzalez, R.; Gauthier, S.A.; Kumar, A.; Levy, E. The exosome secretory pathway transports amyloid precursor protein carboxyl-terminal fragments from the cell into the brain extracellular space. J. Biol. Chem. 2012, 287, 43108–43115. [Google Scholar] [CrossRef]
- Vella, L.J.; Scicluna, B.J.; Cheng, L.; Bawden, E.G.; Masters, C.L.; Ang, C.S.; Willamson, N.; McLean, C.; Barnham, K.J.; Hill, A.F. A rigorous method to enrich for exosomes from brain tissue. J. Extracell. Vesicles 2017, 6, 1348885. [Google Scholar] [CrossRef] [Green Version]
- Hurwitz, S.N.; Sun, L.; Cole, K.Y.; Ford, C.R., 3rd; Olcese, J.M.; Meckes, D.G., Jr. An optimized method for enrichment of whole brain-derived extracellular vesicles reveals insight into neurodegenerative processes in a mouse model of Alzheimer’s disease. J. Neurosci. Methods 2018, 307, 210–220. [Google Scholar] [CrossRef]
- Michaelis, M.L.; Jiang, L.; Michaelis, E.K. Isolation of Synaptosomes, Synaptic Plasma Membranes, and Synaptic Junctional Complexes. Methods Mol. Biol. 2017, 1538, 107–119. [Google Scholar]
- Enright, N.; Simonato, M.; Henshall, D.C. Discovery and validation of blood microRNAs as molecular biomarkers of epilepsy: Ways to close current knowledge gaps. Epilepsia Open 2018, 3, 427–436. [Google Scholar] [CrossRef]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Riwanto, M.; Besler, C.; Knau, A.; Fichtlscherer, S.; Roxe, T.; Zeiher, A.M.; Landmesser, U.; Dimmeler, S. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.M.; Zhao, S.; Ramirez Solano, M.A.; Zhu, W.; Michell, D.L.; Wang, Y.; Shyr, Y.; Sethupathy, P.; Linton, M.F.; Graf, G.A.; et al. Bioinformatic analysis of endogenous and exogenous small RNAs on lipoproteins. J. Extracell. Vesicles 2018, 7, 1506198. [Google Scholar] [CrossRef]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef]
- Tang, Y.T.; Huang, Y.Y.; Zheng, L.; Qin, S.H.; Xu, X.P.; An, T.X.; Xu, Y.; Wu, Y.S.; Hu, X.M.; Ping, B.H.; et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int. J. Mol. Med. 2017, 40, 834–844. [Google Scholar] [CrossRef] [Green Version]
- Rekker, K.; Saare, M.; Roost, A.M.; Kubo, A.L.; Zarovni, N.; Chiesi, A.; Salumets, A.; Peters, M. Comparison of serum exosome isolation methods for microRNA profiling. Clin. Biochem. 2014, 47, 135–138. [Google Scholar] [CrossRef]
- Buschmann, D.; Kirchner, B.; Hermann, S.; Marte, M.; Wurmser, C.; Brandes, F.; Kotschote, S.; Bonin, M.; Steinlein, O.K.; Pfaffl, M.W.; et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles 2018, 7, 1481321. [Google Scholar] [CrossRef]
- Clayton, A.; Buschmann, D.; Byrd, J.B.; Carter, D.R.F.; Cheng, L.; Compton, C.; Daaboul, G.; Devitt, A.; Falcon-Perez, J.; Gardiner, C.; et al. Summary of the ISEV workshop on extracellular vesicles as disease biomarkers, held in Birmingham, UK, during December 2017. J. Extracell. Vesicles 2018, 7, 1473707. [Google Scholar] [CrossRef]
- Lasser, C.; Shelke, G.V.; Yeri, A.; Kim, D.K.; Crescitelli, R.; Raimondo, S.; Sjostrand, M.; Gho, Y.S.; Van Keuren Jensen, K.; Lotvall, J. Two distinct extracellular RNA signatures released by a single cell type identified by microarray and next-generation sequencing. RNA Biol. 2017, 14, 58–72. [Google Scholar] [CrossRef]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Mathivanan, S.; Ji, H.; Simpson, R.J. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol. Cell Proteom. 2013, 12, 587–598. [Google Scholar] [CrossRef]
- Chen, M.; Xu, R.; Ji, H.; Greening, D.W.; Rai, A.; Izumikawa, K.; Ishikawa, H.; Takahashi, N.; Simpson, R.J. Transcriptome and long noncoding RNA sequencing of three extracellular vesicle subtypes released from the human colon cancer LIM1863 cell line. Sci. Rep. 2016, 6, 38397. [Google Scholar] [CrossRef] [Green Version]
- Arraud, N.; Linares, R.; Tan, S.; Gounou, C.; Pasquet, J.M.; Mornet, S.; Brisson, A.R. Extracellular vesicles from blood plasma: Determination of their morphology, size, phenotype and concentration. J. Thromb. Haemost. 2014, 12, 614–627. [Google Scholar] [CrossRef]
- Mustapic, M.; Eitan, E.; Werner, J.K., Jr.; Berkowitz, S.T.; Lazaropoulos, M.P.; Tran, J.; Goetzl, E.J.; Kapogiannis, D. Plasma Extracellular Vesicles Enriched for Neuronal Origin: A Potential Window into Brain Pathologic Processes. Front Neurosci. 2017, 11, 278. [Google Scholar] [CrossRef]
- Faure, J.; Lachenal, G.; Court, M.; Hirrlinger, J.; Chatellard-Causse, C.; Blot, B.; Grange, J.; Schoehn, G.; Goldberg, Y.; Boyer, V.; et al. Exosomes are released by cultured cortical neurones. Mol. Cell. Neurosci. 2006, 31, 642–648. [Google Scholar] [CrossRef]
- Ko, J.; Hemphill, M.A.; Gabrieli, D.; Wu, L.; Yelleswarapu, V.; Lawrence, G.; Pennycooke, W.; Singh, A.; Meaney, D.F.; Issadore, D. Smartphone-enabled optofluidic exosome diagnostic for concussion recovery. Sci. Rep. 2016, 6, 31215. [Google Scholar] [CrossRef]
- Ko, J.; Hemphill, M.; Yang, Z.; Sewell, E.; Na, Y.J.; Sandsmark, D.K.; Haber, M.; Fisher, S.A.; Torre, E.A.; Svane, K.C.; et al. Diagnosis of traumatic brain injury using miRNA signatures in nanomagnetically isolated brain-derived extracellular vesicles. Lab Chip 2018, 18, 3617–3630. [Google Scholar] [CrossRef]
- Kawahara, H.; Hanayama, R. The Role of Exosomes/Extracellular Vesicles in Neural Signal Transduction. Biol. Pharm. Bull. 2018, 41, 1119–1125. [Google Scholar] [CrossRef]
- Fisher, R.S.; Acevedo, C.; Arzimanoglou, A.; Bogacz, A.; Cross, J.H.; Elger, C.E.; Engel, J., Jr.; Forsgren, L.; French, J.A.; Glynn, M.; et al. ILAE official report: A practical clinical definition of epilepsy. Epilepsia 2014, 55, 475–482. [Google Scholar] [CrossRef]
- Scheffer, I.E.; Berkovic, S.; Capovilla, G.; Connolly, M.B.; French, J.; Guilhoto, L.; Hirsch, E.; Jain, S.; Mathern, G.W.; Moshe, S.L.; et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017, 58, 512–521. [Google Scholar] [CrossRef]
- Huttner, H.B.; Corbeil, D.; Thirmeyer, C.; Coras, R.; Kohrmann, M.; Mauer, C.; Kuramatsu, J.B.; Kloska, S.P.; Doerfler, A.; Weigel, D.; et al. Increased membrane shedding-indicated by an elevation of CD133-enriched membrane particles--into the CSF in partial epilepsy. Epilepsy Res. 2012, 99, 101–106. [Google Scholar] [CrossRef]
- Raoof, R.; Jimenez-Mateos, E.M.; Bauer, S.; Tackenberg, B.; Rosenow, F.; Lang, J.; Onugoren, M.D.; Hamer, H.; Huchtemann, T.; Kortvelyessy, P.; et al. Cerebrospinal fluid microRNAs are potential biomarkers of temporal lobe epilepsy and status epilepticus. Sci. Rep. 2017, 7, 3328. [Google Scholar] [CrossRef]
- Raoof, R.; Bauer, S.; El Naggar, H.; Connolly, N.M.C.; Brennan, G.P.; Brindley, E.; Hill, T.; McArdle, H.; Spain, E.; Forster, R.J.; et al. Dual-center, dual-platform microRNA profiling identifies potential plasma biomarkers of adult temporal lobe epilepsy. eBioMedicine 2018, 38, 127–141. [Google Scholar] [CrossRef]
- Yan, S.; Zhang, H.; Xie, W.; Meng, F.; Zhang, K.; Jiang, Y.; Zhang, X.; Zhang, J. Altered microRNA profiles in plasma exosomes from mesial temporal lobe epilepsy with hippocampal sclerosis. Oncotarget 2016, 8, 4136. [Google Scholar] [CrossRef]
- Andrews, A.M.; Lutton, E.M.; Merkel, S.F.; Razmpour, R.; Ramirez, S.H. Mechanical Injury Induces Brain Endothelial-Derived Microvesicle Release: Implications for Cerebral Vascular Injury during Traumatic Brain Injury. Front Cell Neurosci. 2016, 10, 43. [Google Scholar] [CrossRef]
- Kumar, A.; Stoica, B.A.; Loane, D.J.; Yang, M.; Abulwerdi, G.; Khan, N.; Kumar, A.; Thom, S.R.; Faden, A.I. Microglial-derived microparticles mediate neuroinflammation after traumatic brain injury. J. Neuroinflam. 2017, 14, 47. [Google Scholar] [CrossRef]
- Hazelton, I.; Yates, A.; Dale, A.; Roodselaar, J.; Akbar, N.; Ruitenberg, M.J.; Anthony, D.C.; Couch, Y. Exacerbation of Acute Traumatic Brain Injury by Circulating Extracellular Vesicles. J. Neurotrauma 2018, 35, 639–651. [Google Scholar] [CrossRef]
- Bohman, L.E.; Riley, J.; Milovanova, T.N.; Sanborn, M.R.; Thom, S.R.; Armstead, W.M. Microparticles Impair Hypotensive Cerebrovasodilation and Cause Hippocampal Neuronal Cell Injury after Traumatic Brain Injury. J. Neurotrauma 2016, 33, 168–174. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Guo, Y.; Yang, W.; Chen, L.; Ren, D.; Wu, C.; He, B.; Zheng, P.; Tong, W. Phosphorylation of connexin 43 induced by traumatic brain injury promotes exosome release. J. Neurophysiol. 2018, 119, 305–311. [Google Scholar] [CrossRef]
- Midura, E.F.; Jernigan, P.L.; Kuethe, J.W.; Friend, L.A.; Veile, R.; Makley, A.T.; Caldwell, C.C.; Goodman, M.D. Microparticles impact coagulation after traumatic brain injury. J. Surg. Res. 2015, 197, 25–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morel, N.; Morel, O.; Petit, L.; Hugel, B.; Cochard, J.F.; Freyssinet, J.M.; Sztark, F.; Dabadie, P. Generation of procoagulant microparticles in cerebrospinal fluid and peripheral blood after traumatic brain injury. J. Trauma 2008, 64, 698–704. [Google Scholar] [CrossRef]
- Nekludov, M.; Mobarrez, F.; Gryth, D.; Bellander, B.M.; Wallen, H. Formation of microparticles in the injured brain of patients with severe isolated traumatic brain injury. J. Neurotrauma 2014, 31, 1927–1933. [Google Scholar] [CrossRef] [PubMed]
- Patz, S.; Trattnig, C.; Grunbacher, G.; Ebner, B.; Gully, C.; Novak, A.; Rinner, B.; Leitinger, G.; Absenger, M.; Tomescu, O.A.; et al. More than cell dust: Microparticles isolated from cerebrospinal fluid of brain injured patients are messengers carrying mRNAs, miRNAs, and proteins. J. Neurotrauma 2013, 30, 1232–1242. [Google Scholar] [CrossRef] [PubMed]
- Manek, R.; Moghieb, A.; Yang, Z.; Kumar, D.; Kobessiy, F.; Sarkis, G.A.; Raghavan, V.; Wang, K.K.W. Protein Biomarkers and Neuroproteomics Characterization of Microvesicles/Exosomes from Human Cerebrospinal Fluid Following Traumatic Brain Injury. Mol. Neurobiol. 2018, 55, 6112–6128. [Google Scholar] [CrossRef] [PubMed]
- Kuharic, J.; Grabusic, K.; Tokmadzic, V.S.; Stifter, S.; Tulic, K.; Shevchuk, O.; Lucin, P.; Sustic, A. Severe Traumatic Brain Injury Induces Early Changes in the Physical Properties and Protein Composition of Intracranial Extracellular Vesicles. J. Neurotrauma 2018, 36, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. Blymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Szatanek, R.; Baran, J.; Siedlar, M.; Baj-Krzyworzeka, M. Isolation of extracellular vesicles: Determining the correct approach. Int. J. Mol. Med. 2015, 36, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Toffolo, K.; Osei, J.; Kelly, W.; Poulsen, A.; Donahue, K.; Wang, J.; Hunter, M.; Bard, J.; Wang, J.; Poulsen, D. Circulating microRNAs as biomarkers in traumatic brain injury. Neuropharmacology 2019, 145, 199–208. [Google Scholar] [CrossRef]
- Harrison, E.B.; Hochfelder, C.G.; Lamberty, B.G.; Meays, B.M.; Morsey, B.M.; Kelso, M.L.; Fox, H.S.; Yelamanchili, S.V. Traumatic brain injury increases levels of miR-21 in extracellular vesicles: Implications for neuroinflammation. FEBS Open Biol. 2016, 6, 835–846. [Google Scholar] [CrossRef]
- Zhao, R.; Zhou, J.; Dong, X.; Bi, C.; Jiang, R.; Dong, J.; Tian, Y.; Yuan, H.; Zhang, J.N. Circular RNA expression alteration in exosomes from the brain extracellular space after traumatic brain injury in mice. J. Neurotrauma 2018, 35, 2056–2066. [Google Scholar] [CrossRef]
- Zhang, Y.; Chopp, M.; Meng, Y.; Katakowski, M.; Xin, H.; Mahmood, A.; Xiong, Y. Effect of exosomes derived from multipluripotent mesenchymal stromal cells on functional recovery and neurovascular plasticity in rats after traumatic brain injury. J. Neurosurg. 2015, 122, 856–867. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chopp, M.; Zhang, Z.G.; Katakowski, M.; Xin, H.; Qu, C.; Ali, M.; Mahmood, A.; Xiong, Y. Systemic administration of cell-free exosomes generated by human bone marrow derived mesenchymal stem cells cultured under 2D and 3D conditions improves functional recovery in rats after traumatic brain injury. Neurochem. Int. 2016, 111, 69–81. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.Y.; Ren, J.L.; Xu, F.; Chen, F.M.; Li, A. Exosomes secreted by stem cells from human exfoliated deciduous teeth contribute to functional recovery after traumatic brain injury by shifting microglia M1/M2 polarization in rats. Stem Cell Res. Ther. 2017, 8, 198. [Google Scholar] [CrossRef]
- Kim, D.K.; Nishida, H.; An, S.Y.; Shetty, A.K.; Bartosh, T.J.; Prockop, D.J. Chromatographically isolated CD63+CD81+ extracellular vesicles from mesenchymal stromal cells rescue cognitive impairments after TBI. Proc. Natl. Acad. Sci. USA 2016, 113, 170–175. [Google Scholar] [CrossRef]
- Williams, A.M.; Dennahy, I.S.; Bhatti, U.F.; Halaweish, I.; Xiong, Y.; Chang, P.; Nikolian, V.C.; Chtraklin, K.; Brown, J.; Zhang, Y.; et al. Mesenchymal Stem Cell-Derived Exosomes Provide Neuroprotection and Improve Long-Term Neurologic Outcomes in a Swine Model of Traumatic Brain Injury and Hemorrhagic Shock. J. Neurotrauma 2018, 36, 54–60. [Google Scholar] [CrossRef]
- Gao, W.; Li, F.; Liu, L.; Xu, X.; Zhang, B.; Wu, Y.; Yin, D.; Zhou, S.; Sun, D.; Huang, Y.; et al. Endothelial colony-forming cell-derived exosomes restore blood-brain barrier continuity in mice subjected to traumatic brain injury. Exp. Neurol. 2018, 307, 99–108. [Google Scholar] [CrossRef]
- Patel, N.A.; Moss, L.D.; Lee, J.Y.; Tajiri, N.; Acosta, S.; Hudson, C.; Parag, S.; Cooper, D.R.; Borlongan, C.V.; Bickford, P.C. Long noncoding RNA MALAT1 in exosomes drives regenerative function and modulates inflammation-linked networks following traumatic brain injury. J. Neuroinflam. 2018, 15, 204. [Google Scholar] [CrossRef]
- Long, Q.; Upadhya, D.; Hattiangady, B.; Kim, D.K.; An, S.Y.; Shuai, B.; Prockop, D.J.; Shetty, A.K. Intranasal MSC-derived A1-exosomes ease inflammation, and prevent abnormal neurogenesis and memory dysfunction after status epilepticus. Proc. Natl. Acad. Sci. USA 2017, 114, E3536–E3545. [Google Scholar] [CrossRef] [Green Version]
- Rau, T.F.; Kothiwal, A.S.; Rova, A.R.; Brooks, D.M.; Rhoderick, J.F.; Poulsen, A.J.; Hutchinson, J.; Poulsen, D.J. Administration of low dose methamphetamine 12 h after a severe traumatic brain injury prevents neurological dysfunction and cognitive impairment in rats. Exp. Neurol. 2014, 253, 31–40. [Google Scholar] [CrossRef]
- Miszczuk, D.; Debski, K.J.; Tanila, H.; Lukasiuk, K.; Pitkanen, A. Traumatic Brain Injury Increases the Expression of Nos1, Abeta Clearance, and Epileptogenesis in APP/PS1 Mouse Model of Alzheimer’s Disease. Mol. Neurobiol. 2016, 53, 7010–7027. [Google Scholar] [CrossRef]
- Debski, K.J.; Pitkanen, A.; Puhakka, N.; Bot, A.M.; Khurana, I.; Harikrishnan, K.N.; Ziemann, M.; Kaspi, A.; El-Osta, A.; Lukasiuk, K.; et al. Etiology matters—Genomic DNA Methylation Patterns in Three Rat Models of Acquired Epilepsy. Sci. Rep. 2016, 6, 25668. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borras, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [PubMed]
- Volkman, R.; Offen, D. Concise Review: Mesenchymal Stem Cells in Neurodegenerative Diseases. Stem Cells 2017, 35, 1867–1880. [Google Scholar] [CrossRef] [Green Version]
- Costa-Ferro, Z.S.; Vitola, A.S.; Pedroso, M.F.; Cunha, F.B.; Xavier, L.L.; Machado, D.C.; Soares, M.B.; Ribeiro-dos-Santos, R.; DaCosta, J.C. Prevention of seizures and reorganization of hippocampal functions by transplantation of bone marrow cells in the acute phase of experimental epilepsy. Seizure 2010, 19, 84–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa-Ferro, Z.S.; Souza, B.S.; Leal, M.M.; Kaneto, C.M.; Azevedo, C.M.; da Silva, I.C.; Soares, M.B.; Ribeiro-dos-Santos, R.; Dacosta, J.C. Transplantation of bone marrow mononuclear cells decreases seizure incidence, mitigates neuronal loss and modulates pro-inflammatory cytokine production in epileptic rats. Neurobiol. Dis. 2012, 46, 302–313. [Google Scholar] [CrossRef]
- Costa-Ferro, Z.S.; de Borba Cunha, F.; de Freitas Souza, B.S.; Leal, M.M.; da Silva, A.A.; de Bellis Kuhn, T.I.; Forte, A.; Sekiya, E.J.; Soares, M.B.; Dos Santos, R.R. Antiepileptic and neuroprotective effects of human umbilical cord blood mononuclear cells in a pilocarpine-induced epilepsy model. Cytotechnology 2014, 66, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Leal, M.M.; Costa-Ferro, Z.S.; Souza, B.S.; Azevedo, C.M.; Carvalho, T.M.; Kaneto, C.M.; Carvalho, R.H.; Dos Santos, R.R.; Soares, M.B. Early transplantation of bone marrow mononuclear cells promotes neuroprotection and modulation of inflammation after status epilepticus in mice by paracrine mechanisms. Neurochem. Res. 2014, 39, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Fukumura, S.; Sasaki, M.; Kataoka-Sasaki, Y.; Oka, S.; Nakazaki, M.; Nagahama, H.; Morita, T.; Sakai, T.; Tsutsumi, H.; Kocsis, J.D.; et al. Intravenous infusion of mesenchymal stem cells reduces epileptogenesis in a rat model of status epilepticus. Epilepsy Res. 2018, 141, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Liu, Y.; Yan, K.; Chen, L.; Chen, X.R.; Li, P.; Chen, F.F.; Jiang, X.D. Anti-inflammatory and immunomodulatory mechanisms of mesenchymal stem cell transplantation in experimental traumatic brain injury. J. Neuroinflam. 2013, 10, 871. [Google Scholar] [CrossRef]
- Qu, C.; Mahmood, A.; Lu, D.; Goussev, A.; Xiong, Y.; Chopp, M. Treatment of traumatic brain injury in mice with marrow stromal cells. Brain Res. 2008, 1208, 234–239. [Google Scholar] [CrossRef] [Green Version]
- Peng, W.; Sun, J.; Sheng, C.; Wang, Z.; Wang, Y.; Zhang, C.; Fan, R. Systematic review and meta-analysis of efficacy of mesenchymal stem cells on locomotor recovery in animal models of traumatic brain injury. Stem Cell Res. Ther. 2015, 6, 47. [Google Scholar] [CrossRef]
- Drago, D.; Cossetti, C.; Iraci, N.; Gaude, E.; Musco, G.; Bachi, A.; Pluchino, S. The stem cell secretome and its role in brain repair. Biochimie 2013, 95, 2271–2285. [Google Scholar] [CrossRef] [Green Version]
- Pischiutta, F.; Brunelli, L.; Romele, P.; Silini, A.; Sammali, E.; Paracchini, L.; Marchini, S.; Talamini, L.; Bigini, P.; Boncoraglio, G.B.; et al. Protection of Brain Injury by Amniotic Mesenchymal Stromal Cell-Secreted Metabolites. Crit. Care Med. 2016, 44, e1118–e1131. [Google Scholar] [CrossRef]
- Puzar Dominkus, P.; Stenovec, M.; Sitar, S.; Lasic, E.; Zorec, R.; Plemenitas, A.; Zagar, E.; Kreft, M.; Lenassi, M. PKH26 labeling of extracellular vesicles: Characterization and cellular internalization of contaminating PKH26 nanoparticles. Biochim. Biophys. Acta 2018, 1860, 1350–1361. [Google Scholar] [CrossRef]
- Wei, Z.; Batagov, A.O.; Carter, D.R.; Krichevsky, A.M. Fetal Bovine Serum RNA Interferes with the Cell Culture derived Extracellular RNA. Sci. Rep. 2016, 6, 31175. [Google Scholar] [CrossRef] [Green Version]
- Tosar, J.P.; Cayota, A.; Eitan, E.; Halushka, M.K.; Witwer, K.W. Ribonucleic artefacts: Are some extracellular RNA discoveries driven by cell culture medium components? J. Extracell. Vesicles 2017, 6, 1272832. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452. [Google Scholar] [CrossRef]
- Hassel, B.; Tauboll, E.; Shaw, R.; Gjerstad, L.; Dingledine, R. Region-specific changes in gene expression in rat brain after chronic treatment with levetiracetam or phenytoin. Epilepsia 2010, 51, 1714–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]




| Reference | Species | Disease | Tissue | Isolation Method | Analysis Method | Time Point (post-TBI) | Further EV Characterization | Results |
|---|---|---|---|---|---|---|---|---|
| [62] | human | partial epilepsy | CSF | 10,000 g + 200,000 g ultracentrifugation | quantitative immunoblotting | no | no | Larger amount of CD133 on membrane particles in CSF of epilepsy patients |
| [72] | human | severe TBI | CSF | 1500 g + 12,000 g centrifugation | functional prothrombinase assay | 0 d, 3 d, 5 d, 10 d | no | EV number at day 0 post-TBI clearly higher than in control group, but decreased towards day 10 |
| [72] | human | severe TBI | plasma | 1500 g + 12,000 g centrifugation | functional prothrombinase assay | 0 d, 3 d, 5 d, 10 d | no | EV number at day 0 post-TBI higher than in controls, decreased progressively between day 0 and day 10, at day 10 about same level as controls |
| [73] | human | severe TBI | plasma (arterial) | 2000 g + 13,000 g centrifugation | flow cytometry | emergency room, 6 h, 12 h, 24 h, 2 d, 3 d | no | EV number highest at the emergency room, decreased during the 3 days post-TBI |
| [73] | human | severe TBI | plasma (cerebrovenous) | 2000 g + 13,000 g centrifugation | flow cytometry | 6 h, 12 h, 24 h, 2 d, 3 d | no | EV number increased after TBI, decreases during the 3 days post-TBI |
| [74] | human | TBI | CSF | 170,000 g ultracentrifugation | flow cytometry | after TBI | EM and fluorescence microscopy | Increased number of EVs post-TBI |
| [75] | human | severe TBI | CSF | 2 × 100,000 g ultracentrifugation | NTA | 12 h | TEM, Western blot | Increased EV concentration and smaller EVs after TBI |
| [76] | human | severe TBI | CSF | 2 × 100,000 g ultracentrifugation | NTA | d 1, d 2–3, d 4–7 | TEM, Western blot | Highest EV concentration 24 h after injury. Larger EVs on days 4-7 post-TBI |
| [66] | mouse | TBI | plasma | ExoQuick kit | flow cytometry | 24 h | EM | Increased EV number after TBI |
| [67] | mouse | TBI | total blood | 1500 g + 15,000 g + 100,000 g ultracentrifugation | flow cytometry | 24 h | no | Increased number of total blood EVs and microglial EVs after TBI |
| [68] | mouse | TBI | plasma | 120,000 g ultracentrifugation | TRPS | 2 h, 6 h, 12 h, 24 h | TEM, Western blot | Increased EV concentration 24 h after injury. Smaller vesicles at 24 h post-TBI |
| [71] | mouse | TBI | plasma | 10,000 g centrifugation | NTA | 30 min, 3 h, 24 h, 3 d | no | Decreased EV number 3 h and 24 h after injury, returned to sham levels by 3 d post-TBI. The share of platelet-derived (CD41+) EVs increased from 3 h to 24 h. |
| [69] | piglet | TBI | serum | 2300 g centrifugation | flow cytometry | before and after TBI | no | EV number is increased post-TBI |
| [70] | rat | TBI | brain tissue | no isolation | quantitative immunoblotting | 2–8 h | Western blot | Increased expression of CD63 and CD81 in hippocampal EVs at 6 h post-injury. |
| Article | Species | Model | Dose and Time Point | EV Type | What was Measured | Isolation Method | Characterization of EVs | Main Finding |
|---|---|---|---|---|---|---|---|---|
| [82] | rat | controlled cortical impact -induced TBI | 100 µg total proteins, 1 d post-injury | rat MSC EVs | Foot-Fault Test, modified Morris water maze, modified Neurological Severity Score, immunohistochemistry | ExoQuick | Total protein concentration, qNano | EVs improved spatial learning and sensorimotor functional recovery, reduced neuroinflammation and increased the number of newly generated endothelial cells. |
| [83] | rat | controlled cortical impact -induced TBI | 100 µg proteins, 3 × 109 particles, 1 d after injury | human MSC EVs, cultured in 2D and 3D conditions | Modified neurological severity score, foot-fault test, Morris water maze, immunohistochemistry | ExoQuick | Total protein concentration, qNano | EVs enhanced spatial learning, reduced brain inflammation, increased neurogenesis in DG, vascular density and angiogenesis |
| [84] | rat | free -falling method | 100, 250, 500 and 1000 µg/mL, time not mentioned | human exfoliated deciduous teeth stem cell EVs | Basso, Beattie and Bresnahan scores, histopathology and immunofluorescense | ExoQuick | Flow cytometry with CD81, CD63 and CD9, TEM, Western blot with CD9 and CD63 | EVs improved rat motor functional recovery and reduced cortical lesion 2 weeks post-injury |
| [85] | mouse | 1 h post-TBI | human MSC EVs | human MSC EVs | Morris water maze, pattern separation test, immunohistochemistry, cytokines in plasma | Anion exchange column | NTA | EVs rescued pattern separation and spatial learning impairments |
| [86] | swine | computer-controlled cortical impact -induced TBI | 1 × 1013 particles, 9 h, 1 d, 5 d, 9 d, and 13 d post-injury | human MSC EVs | Neurocognitive function test, neurologic severity score (NSS) | Sequential centrifugation | qNano | EV treated animals had better neurological functions first 5 d post-TBI and they completed neurological recovery in shorter time |
| [87] | mouse | controlled cortical impact -induced TBI | EVs from 4 × 106 cells, 2 h post-TBI | endothelial colony-forming cell EVs | Brain water content, beam-walking, corner test, immunofluorescence | Sequential centrifugation | TEM, NTA and Western blot with CD9, CD81 and HSP70 | EVs inhibited PTEN expression, increased AKT expression and reduced Evans blue dye extravasation, brain edema and tight junction degradation |
| [88] | rat | mild controlled cortical impact -induced TBI | 100 µg total proteins, 3 h post-TBI | adipose-derived stem cell EVs | Elevated body swing test, forelimb akinesia, paw grasp, in vivo and ex vivo imaging, immunohistochemistry and RNA sequencing | ExoQuick following magnetic bead capture with CD9, CD63 and CD81 | NTA | MALAT1 containing EVs promoted recovery of function on motor behavior and reduction in cortical brain injury |
| [89] | mouse | pilocarpine-induced SE | 30 µg, approximately 15x10^9 particles, same day and 18 h after SE | human MSC from bone marrow EVs | Object location test, novel object recognition test, pattern separation test, immunostaining, cytokine levels | Anion exchange column | Protein concentration, NTA, anti-inflammatory assay | EVs reduced inflammation in hippocampus, repressed neurodegeneration, aberrant neurogenesis and cognitive and memory impairments |
| Article | Species | Condition | Starting Material | Isolation Method | Characterization of EVs | What was Measured | Main Finding |
|---|---|---|---|---|---|---|---|
| [58] | mouse and human | blast overexposure injury (mice) and TBI patients | plasma and serum | microchip using GluR2 antibody | DLS and SEM in method set-up with CCM | miRNA-seq, 7 miRNAs validated | miRNA-based biomarker panel for diagnosis of TBI |
| [63] | human | TLE and SE patients | CSF | ExoQuick reagent | no characterization | miR-19b-3p, miR-21-5p and miR-451a (top findings) | EV-cargo miRNAs showed more promise than Argonaute2 bound miRNAs as biomarkers |
| [64] | human | TLE patients | plasma | ExoQuick reagent | no characterization | miR-27a, miR-328-3p and miR-654-3p levels (top findings) | Higher diagnostic accuracy with EV-cargo miRNAs as compared to Argonaute2 bound miRNAs |
| [65] | human | mTLE-HS patients | plasma | ExoQuick reagent | TEM or Western blot (not shown) | microarray | 50 differentially expressed miRNAs, 6 validated (miR-3613-5p, miR-4668-5p, miR8071, miR-197-5p, miR-4322 and miR-6781-5p) |
| [74] | human | severe TBI | CSF | ultracentrifugation | TEM, flow cytometry | microarray | 81 miRNAs found, miR-9 and miR-451 differentially packed after TBI |
| [80] | mouse | controlled cortical impact-induced TBI | brain tissue | digestion of brain tissue and ultracentrifugation | TEM | miRNA-seq | miR-212 decreased and miR-21, miR-146, miR-7a, and miR-7b increased |
| [81] | mouse | fluid percussion-induced TBI | extracellular space | digestion of brain tissue and Total Exosome Isolation reagent | TEM, Western blot | circ-RNA-seq | 231 differentially expressed circular RNAs, 5 validated |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karttunen, J.; Heiskanen, M.; Lipponen, A.; Poulsen, D.; Pitkänen, A. Extracellular Vesicles as Diagnostics and Therapeutics for Structural Epilepsies. Int. J. Mol. Sci. 2019, 20, 1259. https://doi.org/10.3390/ijms20061259
Karttunen J, Heiskanen M, Lipponen A, Poulsen D, Pitkänen A. Extracellular Vesicles as Diagnostics and Therapeutics for Structural Epilepsies. International Journal of Molecular Sciences. 2019; 20(6):1259. https://doi.org/10.3390/ijms20061259
Chicago/Turabian StyleKarttunen, Jenni, Mette Heiskanen, Anssi Lipponen, David Poulsen, and Asla Pitkänen. 2019. "Extracellular Vesicles as Diagnostics and Therapeutics for Structural Epilepsies" International Journal of Molecular Sciences 20, no. 6: 1259. https://doi.org/10.3390/ijms20061259
APA StyleKarttunen, J., Heiskanen, M., Lipponen, A., Poulsen, D., & Pitkänen, A. (2019). Extracellular Vesicles as Diagnostics and Therapeutics for Structural Epilepsies. International Journal of Molecular Sciences, 20(6), 1259. https://doi.org/10.3390/ijms20061259