Genome-Wide Analysis of LIM Family Genes in Foxtail Millet (Setaria italica L.) and Characterization of the Role of SiWLIM2b in Drought Tolerance
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Analysis and Chromosomal Distribution of 10 LIM-Like Genes
2.2. Functional Domains and Gene Structure Analysis of the 10 LIM-Like Genes
2.3. Promoter Analysis and Tissue-Specific Expression of the LIM Genes in Foxtail Millet
2.4. Analysis of SiWLIM2b Expression under Various Treatments
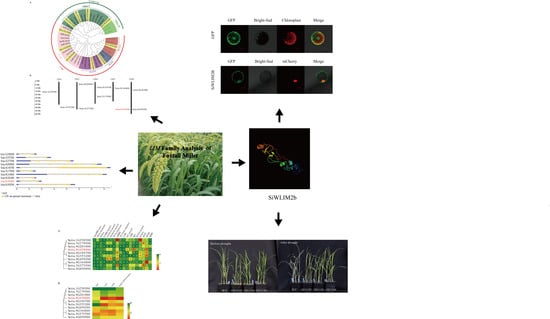
2.5. Subcellular Location of the SiWLIM2b Protein
2.6. Overexpression of SiWLIM2b Enhances Drought Resistancein Transgenic Rice
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification and Phylogenetic Analysis of LIMFamily Genes
4.3. Chromosome Locations and Gene Structures of LIM Family Genes and Identification of Functional Domains and Cis-Acting Elements
4.4. Tissue-Specific Expression Profiling Using RNA-Seq Data
4.5. Total RNA Extraction and qRT-PCR Analysis
4.6. Subcellular Localization of SiWLIM2b
4.7. Phenotypic Analysis of Transgenic Rice in a Greenhouse
4.8. Differential Expression of Phenylpropane Secondary Metabolic Pathway Genes in Transgenic and Control Rice Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PAL | Phe ammonia lyase |
| ROS | Reactive oxygen species |
| MDA | malondialdehyde |
| 4CL2 | 4-coumarate coenzyme A (CoA) ligase 2 |
| C3H | Coumarate-3-hydroxylase |
| SA | salicylic acid |
| MeJA | methyl jasmonate |
| GA | gibberellic acid |
| ABRE | ABA-responsive element |
| qRT-PCR | quantitative real-time PCR |
| GFP | green fluorescent protein |
| ND | nitrogen deficiency |
| ABA | abscisic acid |
References
- Eliasson, A.; Gass, N.; Mundel, C.; Baltz, R.; Krauter, R.; Evrard, J.L.; Steinmetz, A. Molecular and expression analysis of a lim protein gene family from flowering plants. Mol. Gen. Genet. 2000, 264, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Kadrmas, J.L.; Beckerle, M.C. The lim domain: From the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 2004, 5, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.; Verma, P.K. The plant lim proteins: Unlocking the hidden attractions. Planta 2017, 246, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.J.; Wan, A.R.; Jauh, G.Y. An actin-binding protein, lllim1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 2008, 147, 1619–1636. [Google Scholar] [CrossRef] [PubMed]
- Kawaoka, A.; Ebinuma, H. Transcriptional control of lignin biosynthesis by tobacco lim protein. Phytochemistry 2001, 57, 1149–1157. [Google Scholar] [CrossRef]
- Kawaoka, A.; Kaothien, P.; Yoshida, K.; Endo, S.; Yamada, K.; Ebinuma, H. Functional analysis of tobacco lim protein ntlim1 involved in lignin biosynthesis. Plant J. Cell Mol. Biol. 2000, 22, 289–301. [Google Scholar] [CrossRef]
- Labalette, C.; Nouet, Y.; Levillayer, F.; Colnot, S.; Chen, J.; Claude, V.; Huerre, M.; Perret, C.; Buendia, M.A.; Wei, Y. Deficiency of the lim-only protein fhl2 reduces intestinal tumorigenesis in apc mutant mice. PLoS ONE 2010, 5, e10371. [Google Scholar] [CrossRef] [PubMed]
- Way, J.C.; Chalfie, M. Mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in c. Elegans. Cell 1988, 54, 5–16. [Google Scholar] [CrossRef]
- Freyd, G.; Kim, S.K.; Horvitz, H.R. Novel cysteine-rich motif and homeodomain in the product of the caenorhabditis elegans cell lineage gene lin-11. Nature 1990, 344, 876–879. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, O.; Thor, S.; Norberg, T.; Ohlsson, H.; Edlund, T. Insulin gene enhancer binding protein isl-1 is a member of a novel class of proteins containing both a homeo- and a cys-his domain. Nature 1990, 344, 879–882. [Google Scholar] [CrossRef]
- Dawid, I.B.; Toyama, R.; Taira, M. Lim domain proteins. C. R. l’Acad. Sci. Ser. III Sci. 1995, 318, 295–306. [Google Scholar]
- Perez-Alvarado, G.C.; Miles, C.; Michelsen, J.W.; Louis, H.A.; Winge, D.R.; Beckerle, M.C.; Summers, M.F. Structure of the carboxy-terminal lim domain from the cysteine rich protein crp. Nat. Struct. Biol. 1994, 1, 388–398. [Google Scholar] [CrossRef]
- Baltz, R.; Evrard, J.L.; Bourdon, V.; Steinmetz, A. The pollen-specific lim protein plim-1 from sunflower binds nucleic acids in vitro. Sex. Plant Reprod. 1996, 9, 264–268. [Google Scholar] [CrossRef]
- Moes, D.; Gatti, S.; Hoffmann, C.; Dieterle, M.; Moreau, F.; Neumann, K.; Schumacher, M.; Diederich, M.; Grill, E.; Shen, W.H.; et al. A lim domain protein from tobacco involved in actin-bundling and histone gene transcription. Mol. Plant 2013, 6, 483–502. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Hoffmann, C.; Dieterle, M.; Van Troys, M.; Ampe, C.; Steinmetza, A. Tobacco wlim1 is a novel f-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 2006, 18, 2194–2206. [Google Scholar] [CrossRef]
- Papuga, J.; Thomas, C.; Dieterle, M.; Moreau, F.; Steinmetz, A. Arabidopsis lim domain proteins involved in actin bundling exhibit different modes of regulation. FEBS J. 2009, 276, 245. [Google Scholar]
- Papuga, J.; Hoffmann, C.; Dieterle, M.; Moes, D.; Moreau, F.; Tholl, S.; Steinmetz, A.; Thomas, C. Arabidopsis lim proteins: A family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell 2010, 22, 3034–3052. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, D.; Dejardin, A.; Leple, J.C.; Lesage-Descauses, M.C.; Pilate, G. Genome-wide analysis of lim gene family in populus trichocarpa, arabidopsis thaliana, and oryza sativa. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2007, 14, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, J.; Li, L.; Wang, X.L.; Wang, N.N.; Li, D.D.; Li, X.B. A cotton lim domain-containing protein (ghwlim5) is involved in bundling actin filaments. Plant Physiol. Biochem. 2013, 66, 34–40. [Google Scholar] [CrossRef]
- Li, L.; Li, Y.; Wang, N.N.; Li, Y.; Lu, R.; Li, X.B. Cotton lim domain-containing protein ghplim1 is specifically expressed in anthers and participates in modulating f-actin. Plant Biol. 2015, 17, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Han, L.B.; Li, Y.B.; Wang, H.Y.; Wu, X.M.; Li, C.L.; Luo, M.; Wu, S.J.; Kong, Z.S.; Pei, Y.; Jiao, G.L.; et al. The dual functions of wlim1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell 2013, 25, 4421–4438. [Google Scholar] [CrossRef] [PubMed]
- Khatun, K.; Robin, A.H.K.; Park, J.I.; Ahmed, N.U.; Kim, C.K.; Lim, K.B.; Kim, M.B.; Lee, D.J.; Nou, I.S.; Chung, M.Y. Genome-wide identification, characterization and expression profiling of lim family genes in solanum lycopersicum L. Plant Physiol. Biochem. 2016, 108, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; He, L.; Gu, Y.; Wang, Y.; Chen, Q.; He, C. Genome-wide analyses of a plant-specific lim-domain gene family implicate its evolutionary role in plant diversification. Genome Biol. Evol. 2014, 6, 1000–1012. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant. 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Fraser, C.M.; Chapple, C. The phenylpropanoid pathway in arabidopsis. Arabidopsis Book 2011, 9, e0152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, C.J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 2015, 8, 17–27. [Google Scholar] [CrossRef]
- Sala, S.; Ampe, C. An emerging link between lim domain proteins and nuclear receptors. Cell. Mol. Life Sci. 2018, 75, 1959–1971. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Zhao, Y. The diverse biofunctions of lim domain proteins: Determined by subcellular localization and protein-protein interaction. Biol. Cell 2007, 99, 489–502. [Google Scholar] [CrossRef]
- Kaothien, P.; Kawaoka, A.; Ebinuma, H.; Yoshida, K.; Shinmyo, A. Ntlim1, a pal-box binding factor, controls promoter activity of the horseradish wound-inducible peroxidase gene. Plant Mol. Biol. 2002, 49, 591–599. [Google Scholar] [CrossRef]
- Cheng, X.; Li, G.; Muhammad, A.; Zhang, J.; Jiang, T.; Jin, Q.; Zhao, H.; Cai, Y.; Lin, Y. Molecular identification, phylogenomic characterization and expression patterns analysis of the lim (lin-11, isl1 and mec-3 domains) gene family in pear (pyrus bretschneideri) reveal its potential role in lignin metabolism. Gene 2018. [Google Scholar] [CrossRef] [PubMed]
- Wasteneys, G.O.; Yang, Z. The cytoskeleton becomes multidisciplinary. Plant Physiol. 2004, 136, 3853–3854. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, L.J.; Huang, R.D. Cytoskeleton and plant salt stress tolerance. Plant Signal. Behav. 2011, 6, 29–31. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Valliyodan, B.; Zhang, J.; Lenoble, M.E.; Yu, O.; Rogers, E.E.; Nguyen, H.T.; Sharp, R.E. Regulation of growth response to water stress in the soybean primary root. I. Proteomic analysis reveals region-specific regulation of phenylpropanoid metabolism and control of free iron in the elongation zone. Plant Cell Environ. 2010, 33, 223–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.J.; Xiong, L.Z. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Lata, C.; Gupta, S.; Prasad, M. Foxtail millet: A model crop for genetic and genomic studies in bioenergy grasses. Crit. Rev. Biotechnol. 2013, 33, 328–343. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.F.; Zhao, W.Y.; Fu, J.D.; Liu, Y.W.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; Xi, Y.J. Genome-wide analysis of cdpk family in foxtail millet and determination of sicdpk24 functions in drought stress. Front. Plant Sci. 2018, 9, 651. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, X.; Quan, Z.; Cheng, S.; Xu, X.; Pan, S.; Xie, M.; Zeng, P.; Yue, Z.; Wang, W.; et al. Genome sequence of foxtail millet (setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 2012, 30, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Bennetzen, J.L.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.C.; Estep, M.; Feng, L.; Vaughn, J.N.; Grimwood, J.; et al. Reference genome sequence of the model plant setaria. Nat. Biotechnol. 2012, 30, 555–561. [Google Scholar] [CrossRef]
- Jiangtao, C.; Yingzhen, K.; Qian, W.; Yuhe, S.; Daping, G.; Jing, L.; Guanshan, L. Mapgene2chrom, a tool to draw gene physical map based on perl and svg languages. Yi Chuan (Hereditas) 2015, 37, 91–97. [Google Scholar]
- Zhang, Y.; Mao, L.; Wang, H.; Brocker, C.; Yin, X.; Vasiliou, V.; Fei, Z.; Wang, X. Genome-wide identification and analysis of grape aldehyde dehydrogenase (aldh) gene superfamily. PLoS ONE 2012, 7, e32153. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011, 13, 59–69. [Google Scholar] [CrossRef]
- Wong, D.C.J.; Lopez Gutierrez, R.; Gambetta, G.A.; Castellarin, S.D. Genome-wide analysis of cis-regulatory element structure and discovery of motif-driven gene co-expression networks in grapevine. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2017, 24, 311–326. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.F.; Schraut, D.; Hartung, W.; Schaffner, A.R. Differential responses of maize mip genes to salt stress and aba. J. Exp. Bot. 2005, 56, 2971–2981. [Google Scholar] [CrossRef]
- Faraji, S.; Rasouli, S.H.; Kazemitabar, S.K. Genome-wide exploration of c2h2 zinc finger family in durum wheat (triticum turgidum ssp durum): Insights into the roles in biological processes especially stress response. Biometals 2018, 31, 1019–1042. [Google Scholar] [CrossRef]
- Onishi, M.; Tachi, H.; Kojima, T.; Shiraiwa, M.; Takahara, H. Molecular cloning and characterization of a novel salt-inducible gene encoding an acidic isoform of pr-5 protein in soybean (glycine max [L.] merr.). Plant Physiol. Biochem. 2006, 44, 574–580. [Google Scholar] [CrossRef]
- Cochrane, G.; Alako, B.; Amid, C.; Bower, L.; Cerdeno-Tarraga, A.; Cleland, I.; Gibson, R.; Goodgame, N.; Jang, M.; Kay, S.; et al. Facing growth in the european nucleotide archive. Nucleic Acids Res. 2013, 41, D30–D35. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Bonthala, V.S.; Mishra, A.K.; Khandelwal, R.; Khan, Y.; Roy, R.; Prasad, M. C2h2 type of zinc finger transcription factors in foxtail millet define response to abiotic stresses. Funct. Integr. Genom. 2014, 14, 531–543. [Google Scholar] [CrossRef]
- Miller, M.J.; Barrett-Wilt, G.A.; Hua, Z.; Vierstra, R.D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 16512–16517. [Google Scholar] [CrossRef]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and characterization of gmmyb118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef]
- Liu, J.Y.; Osbourn, A.; Ma, P.D. Myb transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Shen, X.X.; Salichos, L.; Rokas, A. A genome-scale investigation of how sequence, function, and tree-based gene properties influence phylogenetic inference. Genome Biol. Evol. 2016, 8, 2565–2580. [Google Scholar] [CrossRef]
- Kellogg, E.A. Evolutionary history of the grasses. Plant Physiol. 2001, 125, 1198–1205. [Google Scholar] [CrossRef]
- Shaul, O. How introns enhance gene expression. Int. J. Biochem. Cell Biol. 2017, 91, 145–155. [Google Scholar] [CrossRef]
- Yan, N.; Doelling, J.H.; Falbel, T.G.; Durski, A.M.; Vierstra, R.D. The ubiquitin-specific protease family from arabidopsis. Atubp1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 2000, 124, 1828–1843. [Google Scholar] [CrossRef]
- Raasi, S.; Wolf, D.H. Ubiquitin receptors and erad: A network of pathways to the proteasome. Semin. Cell Dev. Biol. 2007, 18, 780–791. [Google Scholar] [CrossRef]
- King, R.W.; Deshaies, R.J.; Peters, J.M.; Kirschner, M.W. How proteolysis drives the cell cycle. Science 1996, 274, 1652–1659. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, L.; Corke, F.; Smith, C.; Bevan, M.W. Control of final seed and organ size by the da1 gene family in arabidopsis thaliana. Genes Dev. 2008, 22, 1331–1336. [Google Scholar] [CrossRef]
- Xia, T.; Li, N.; Dumenil, J.; Li, J.; Kamenski, A.; Bevan, M.W.; Gao, F.; Li, Y. The ubiquitin receptor da1 interacts with the e3 ubiquitin ligase da2 to regulate seed and organ size in arabidopsis. Plant Cell 2013, 25, 3347–3359. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, Y.W.; Zhou, J.M.; Zhao, S.P.; Zhang, X.H.; Min, D.H. Genome-wide analysis of the lectin receptor-like kinase family in foxtail millet (setaria italica L.). Plant Cell Tissue Organ Culture 2016, 127, 335–346. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; van Wees, S.C.M.; van Pelt, J.A.; Knoester, M.; Laan, R.; Gerrits, N.; Weisbeek, P.J.; van Loon, L.C. A novel signaling pathway controlling induced systemic resistance in arabidopsis. Plant Cell 1998, 10, 1571–1580. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, G.J.; Yan, X.F.; Wei, Z.G.; Xu, Z.R. Meja-inducible expression of the heterologous jaz2 promoter from arabidopsis in populus trichocarpa protoplasts. J. Plant Dis. Prot. 2011, 118, 69–74. [Google Scholar] [CrossRef]
- Kim, J.S.; Mizoi, J.; Yoshida, T.; Fujita, Y.; Nakajima, J.; Ohori, T.; Todaka, D.; Nakashima, K.; Hirayama, T.; Shinozaki, K.; et al. An abre promoter sequence is involved in osmotic stress-responsive expression of the dreb2a gene, which encodes a transcription factor regulating drought-inducible genes in arabidopsis. Plant Cell Physiol. 2011, 52, 2136–2146. [Google Scholar] [CrossRef]
- Park, J.I.; Ahmed, N.U.; Jung, H.J.; Arasan, S.K.; Chung, M.Y.; Cho, Y.G.; Watanabe, M.; Nou, I.S. Identification and characterization of lim gene family in brassica rapa. BMC Genom. 2014, 15, 641. [Google Scholar] [CrossRef]
- Gawel, S.; Wardas, M.; Niedworok, E.; Wardas, P. [malondialdehyde (mda) as a lipid peroxidation marker]. Wiadomosci Lekarskie 2004, 57, 453–455. [Google Scholar]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (mda) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef]
- Karatas, I.; Ozturk, L.; Demir, Y.; Unlukara, A.; Kurunc, A.; Duzdemir, O. Alterations in antioxidant enzyme activities and proline content in pea leaves under long-term drought stress. Toxicol. Ind. Health 2014, 30, 693–700. [Google Scholar] [CrossRef]
- Wei, Q.; Zhang, F.; Sun, F.; Luo, Q.; Wang, R.; Hu, R.; Chen, M.; Chang, J.; Yang, G.; He, G. A wheat myb transcriptional repressor tamyb1d regulates phenylpropanoid metabolism and enhances tolerance to drought and oxidative stresses in transgenic tobacco plants. Plant Sci. Int. J. Exp. Plant Biol. 2017, 265, 112–123. [Google Scholar] [CrossRef]
- Mahapatro, G.; Mishra, D.; Shaw, K.; Mishra, S.; Jena, T. Phylogenetic tree construction for DNA sequences using clustering methods. Procedia Eng. 2012, 38, 1362–1366. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. Mega: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. Gsds 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- He, Z.L.; Zhang, H.K.; Gao, S.H.; Lercher, M.J.; Chen, W.H.; Hu, S.N. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Jungklang, J.; Saengnil, K.; Uthaibutra, J. Effects of water-deficit stress and paclobutrazol on growth, relative water content, electrolyte leakage, proline content and some antioxidant changes in curcuma alismatifolia gagnep. Cv. Chiang mai pink. Saudi J. Biol. Sci. 2017, 24, 1505–1512. [Google Scholar] [CrossRef]
- Smart, R.E. Rapid estimates of relative water content. Plant Physiol. 1974, 53, 258–260. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, R.; Chen, M.; Sun, J.-C.; Yu, Y.; Min, D.-H.; Chen, J.; Xu, Z.-S.; Zhou, Y.-B.; Ma, Y.-Z.; Zhang, X.-H. Genome-Wide Analysis of LIM Family Genes in Foxtail Millet (Setaria italica L.) and Characterization of the Role of SiWLIM2b in Drought Tolerance. Int. J. Mol. Sci. 2019, 20, 1303. https://doi.org/10.3390/ijms20061303
Yang R, Chen M, Sun J-C, Yu Y, Min D-H, Chen J, Xu Z-S, Zhou Y-B, Ma Y-Z, Zhang X-H. Genome-Wide Analysis of LIM Family Genes in Foxtail Millet (Setaria italica L.) and Characterization of the Role of SiWLIM2b in Drought Tolerance. International Journal of Molecular Sciences. 2019; 20(6):1303. https://doi.org/10.3390/ijms20061303
Chicago/Turabian StyleYang, Rui, Ming Chen, Jian-Chang Sun, Yue Yu, Dong-Hong Min, Jun Chen, Zhao-Shi Xu, Yong-Bin Zhou, You-Zhi Ma, and Xiao-Hong Zhang. 2019. "Genome-Wide Analysis of LIM Family Genes in Foxtail Millet (Setaria italica L.) and Characterization of the Role of SiWLIM2b in Drought Tolerance" International Journal of Molecular Sciences 20, no. 6: 1303. https://doi.org/10.3390/ijms20061303
APA StyleYang, R., Chen, M., Sun, J. -C., Yu, Y., Min, D. -H., Chen, J., Xu, Z. -S., Zhou, Y. -B., Ma, Y. -Z., & Zhang, X. -H. (2019). Genome-Wide Analysis of LIM Family Genes in Foxtail Millet (Setaria italica L.) and Characterization of the Role of SiWLIM2b in Drought Tolerance. International Journal of Molecular Sciences, 20(6), 1303. https://doi.org/10.3390/ijms20061303