BcXyl, a β-xylosidase Isolated from Brunfelsia Calycina Flowers with Anthocyanin-β-glycosidase Activity
Abstract
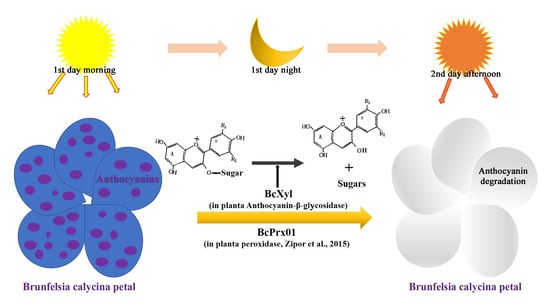
:1. Introduction
2. Results
2.1. High Level of β-Glycosidase Activity in Brunfelsia Calycina Flower Petals during Development
2.2. A β-Glycosidase was Purified from B. calycina Petals
2.3. The Purified β-Glycosidase was a β-Xylosidase
2.4. Enzymatic Properties of the β-Xylosidase (BcXyl)
2.5. BcXyl Showed Glycosidase Activity to the Highly Purified Anthocyanins from B. calycina
2.6. Full-length BcXyl mRNA Sequence and Its Abundance during Flower Development
3. Discussion
3.1. A β-Xylosidase was Purified from B. calycina Flower and Proved to Catalyze the Hydrolysis of the Glycosidic Bonds in Anthocyanins
3.2. BcXyl Belongs to Glycoside Hydrolase Family 3 as Two Fungal Characterized Anthocyanin-β-Glycosidases
3.3. The Role of BcXyl in the Rapid Degradation of Petal Anthocyanins
4. Materials and Methods
4.1. Plant Materials
4.2. Anthocyanin Content Determination
4.3. Crude Enzyme Extraction, protein content and β-Glycosidase Activity Assay
4.4. Purification and Identification of a β-Xylosidase in B. calycina Petals
4.5. Effect of pH and Temperature on Enzyme Activity
4.6. Influence of Metal Ion and Chemical Effectors on Enzyme Activity
4.7. Substrate Specificity
4.8. Anthocyanin Purification
4.9. High Performance Liquid Chromatography (HPLC) Analysis of Anthocyanin Degradation by Brunfelsia β-Xylosidase (BcXyl)
4.10. Identification and Quantification of Sugars Released from Anthocyanin Degradation by BcXyl by Gas Chromatography-Mass Spectrometry
4.11. BcXyl cDNA Cloning, Sequence Features, Potential 3D Structure Prediction of the Deduced Protein Sequence
4.12. Molecular Phylogenetic Analysis of BcXyl
4.13. BcXyl Transcript Abundance Analysis
4.14. Statistics Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Ohmiya, A. Seeing is believing: Engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotechnol. 2008, 19, 190–197. [Google Scholar] [CrossRef]
- Passeri, V.; Koes, R.; Quattrocchio, F.M. New challenges for the design of high value plant products: Stabilization of anthocyanins in plant vacuoles. Front. Plant Sci. 2016, 7, 153. [Google Scholar] [CrossRef]
- Oren-Shamir, M. Does anthocyanin degradation play a significant role in determining pigment concentration in plants? Plant Sci. 2009, 177, 310–316. [Google Scholar] [CrossRef]
- Vaknin, H.; Bar-Akiva, A.; Ovadia, R.; Nissim-Levi, A.; Forer, I.; Weiss, D.; Oren-Shamir, M. Active anthocyanin degradation in Brunfelsia calycina (yesterday–today–tomorrow) flowers. Planta 2005, 222, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Schmitzer, V.; Veberic, R.; Osterc, G.; Stampar, F. Changes in the phenolic concentration during flower development of rose ‘KORcrisett’. J. Am. Soc. Hortic. Sci. 2009, 134, 491–496. [Google Scholar] [CrossRef]
- Steyn, W.J.; Holcroft, D.M.; Wand, S.J.E.; Jacobs, G. Anthocyanin degradation in detached pome fruit with reference to preharvest red color loss and pigmentation patterns of blushed and fully red pears. J. Am. Soc. Hortic. Sci. 2004, 129, 13–19. [Google Scholar] [CrossRef]
- Zhang, Z.; Pang, X.; Ji, Z.; Jiang, Y. Role of anthocyanin degradation in litchi pericarp browning. Food Chem. 2001, 75, 217–221. [Google Scholar] [CrossRef]
- Zhang, Z.; Pang, X.; Duan, X.; Ji, Z.; Jiang, Y. Role of peroxidase in anthocyanin degradation in litchi fruit pericarp. Food Chem. 2005, 90, 47–52. [Google Scholar] [CrossRef]
- Barbagallo, R.N.; Palmeri, R.; Fabiano, S.; Rapisarda, P.; Spagna, G. Characteristic of β-glucosidase from Sicilian blood oranges in relation to anthocyanin degradation. Enzyme Microb. Technol. 2007, 41, 570–575. [Google Scholar] [CrossRef]
- Fang, F.; Zhang, X.; Luo, H.; Zhou, J.; Gong, Y.; Li, W.; Shi, Z.; He, Q.; Wu, Q.; Li, L.; et al. An intracellular laccase is responsible for epicatechin-mediated anthocyanin degradation in litchi fruit pericarp. Plant Physiol. 2015, 169, 2391–2408. [Google Scholar] [CrossRef]
- Zipor, G.; Duarte, P.; Carqueijeiro, I.; Shahar, L.; Ovadia, R.; Teper-Bamnolker, P.; Eshel, D.; Levin, Y.; Doron-Faigenboim, A.; Sottomayor, M.; et al. In planta anthocyanin degradation by a vacuolar class III peroxidase in Brunfelsia calycina flowers. New Phytol. 2015, 205, 653–665. [Google Scholar] [CrossRef]
- Suzuki, H.; Takahashi, S.; Watanabe, R.; Fukushima, Y.; Fujita, N.; Noguchi, A.; Yokoyama, R.; Nishitani, K.; Nishino, T.; Nakayama, T. An isoflavone conjugate-hydrolyzing β-Glucosidase from the roots of soybean (Glycine max) seedlings purification, gene cloning, phylogenetics, and cellular localization. J. Biol. Chem. 2006, 281, 30251–30259. [Google Scholar] [CrossRef]
- Rivera-Vargas, L.I.; Schmitthenner, A.F.; Graham, T.L. Soybean flavonoid effects on and metabolism by Phytophthora sojae. Phytochemistry 1993, 32, 851–857. [Google Scholar] [CrossRef]
- Dakora, F.; Phillips, D.A. Diverse functions of isoflavonoids in legumes transcend anti-microbial definitions of phytoalexins. Physiol. Mol. Plant Pathol. 1996, 49, 1–20. [Google Scholar] [CrossRef]
- Graham, T.L.; Graham, M.Y. Signaling in soybean phenylpropanoid responses (dissection of primary, secondary, and conditioning effects of light, wounding, and elicitor treatments). Plant Physiol. 1996, 110, 1123–1133. [Google Scholar] [CrossRef]
- Manzanares, P.; Rojas, V.; Genovés, S.; Vallés, S. A preliminary search for anthocyanin-β-d-glucosidase activity in non-Saccharomyces wine yeasts. Int. J. Food Sci. Technol. 2000, 35, 95–103. [Google Scholar] [CrossRef]
- Blom, H. Partial characterization of a thermostable anthocyanin-β-glycosidase from Aspergillus niger. Food Chem. 1983, 12, 197–204. [Google Scholar] [CrossRef]
- Sánchez-Torres, P.; González-Candelas, L.; Ramón, D. Heterologous expression of a Candida molischiana anthocyanin-β-glucosidase in a wine yeast strain. J. Agric. Food Chem. 1998, 46, 354–360. [Google Scholar] [CrossRef]
- Bar-Akiva, A.; Ovadia, R.; Rogachev, I.; Bar-Or, C.; Bar, E.; Freiman, Z.; Nissim-Levi, A.; Gollop, N.; Lewinsohn, E.; Aharoni, A.; et al. Metabolic networking in Brunfelsia calycina petals after flower opening. J. Exp. Bot. 2010, 61, 1393–1403. [Google Scholar] [CrossRef]
- Cournoyer, B.; Faure, D. Radiation and functional specialization of the family-3 glycoside hydrolases. J. Mol. Microbiol. Biotechnol. 2003, 5, 190–198. [Google Scholar] [CrossRef]
- Behrens, C.J.; Krahe, N.K.; Linke, D.; Berger, R.G. BadGluc, a β-glucosidase from Bjerkandera adusta with anthocyanase properties. Bioprocess Biosyst. Eng. 2018, 41, 1391–1401. [Google Scholar] [CrossRef]
- Sakamura, S.; Obata, Y. Anthocyanase and Anthocyanins Occurring in Eggplant, Solanum melongena L.(I). Agric. Biol. Chem. 1961, 25, 750–756. [Google Scholar] [CrossRef]
- Janbon, G.; Derancourt, J.; Chemardin, P.; Arnaud, A.; Galzy, P. A very stable β-glucosidase from a Candida molischiana mutant strain: Enzymatic properties, sequencing, and homology with other yeast β-glucosidases. Biosci. Biotechnol. Biochem. 1995, 59, 1320–1322. [Google Scholar] [CrossRef]
- Janbon, G.; Magnet, R.; Arnaud, A.; Galzy, P. Cloning and sequencing of the β-glucosidase-encoding gene from Candida molischiana strain 35M5N. Gene 1995, 165, 109–113. [Google Scholar] [CrossRef]
- Hósel, W.; Barz, W. β-Glucosidases from Cicer arietinum L. Purification and Properties of Isoflavone-7-O-glucoside-Specific β-Glucosidases. Eur. J. Biochem. 1975, 57, 607–616. [Google Scholar] [CrossRef]
- Svasti, J.; Srisomsap, C.; Techasakul, S.; Surarit, R. Dalcochinin-8′-O-β-d-glucoside and its β-glucosidase enzyme from Dalbergia cochinchinensis. Phytochemistry 1999, 50, 739–743. [Google Scholar] [CrossRef]
- Chuankhayan, P.; Rimlumduan, T.; Svasti, J.; Cairns, J.R. Hydrolysis of soybean isoflavonoid glycosides by Dalbergia β-glucosidases. J. Agric. Food Chem. 2007, 55, 2407–2412. [Google Scholar] [CrossRef]
- Castañeda-Ovando, A.; Pacheco-Hernández, M.L.; Páez-Hernández, M.E.; Rodríguez, J.A.; Galán-Vidal, C.A. Chemical studies of anthocyanins: A review. Food Chem. 2009, 113, 859–871. [Google Scholar] [CrossRef]
- Luo, H.; Deng, S.; Fu, W.; Zhang, X.; Zhang, X.; Zhang, Z.; Pang, X. Characterization of active anthocyanin degradation in the petals of rosa chinensis and brunfelsia calycina reveals the effect of gallated catechins on pigment maintenance. Int. J. Mol. Sci. 2017, 18, 699. [Google Scholar] [CrossRef]
- Li, N.; Han, X.; Xu, S.; Li, C.; Wei, X.; Liu, Y.; Zhang, R.; Tang, X.; Zhou, J.; Huang, Z. Glycoside Hydrolase Family 39 β-Xylosidase of Sphingomonas Showing Salt/Ethanol/Trypsin Tolerance, Low-pH/Low-Temperature Activity, and Transxylosylation Activity. J. Agric. Food Chem. 2018, 66, 9465–9472. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Zhang, Z.; Pang, X.; Yang, C.; Ji, Z.; Jiang, Y. Purification and structural analysis of anthocyanins from litchi pericarp. Food Chem. 2004, 84, 601–604. [Google Scholar] [CrossRef]
- Martinez, V.; Ingwers, M.; Smith, J.; Glushka, J.; Yang, T.; Bar-Peled, M. Biosynthesis of UDP-4-keto-6-deoxyglucose and UDP-rhamnose in pathogenic fungi Magnaporthe grisea and Botryotinia fuckeliana. J. Biol. Chem. 2012, 287, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]






| Purification Step | Total Activity (μmol/h) 1 | Total Protein (mg) | Specific Activity (μmol/h/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| crude extract | 614.64 | 85.93 | 7.15 | 1.00 | 100.00 |
| (NH4)2SO4 | 295.74 | 13.96 | 21.19 | 2.96 | 48.12 |
| DEAE Sepharose 2 | 136.89 | 3.68 | 37.23 | 5.21 | 22.27 |
| CM-Sepharose 3 | 22.30 | 0.07 | 318.57 | 44.56 | 3.63 |
| Sequences Detected by MS/MS | Annotation | Gene Identified | Organisms | |
|---|---|---|---|---|
| 1 | AVSNNFATLMR | beta-xylosidase/alpha-l-arabinofuranosidase | XP_009612011.1; XP_006351808.1; XP_009762535.1 | Nicotiana tomentosiformis; Solanum tuberosum; Nicotiana sylvestris |
| 2 | LPMTWYPQSYADK | beta-xylosidase /alpha-l-arabinofuranosidase | XP_009612011.1; XP_009762535.1 | Nicotiana tomentosiformis; Nicotiana sylvestris |
| 3 | VTQQDLDDTFNPPFK | beta-xylosidase /alpha-l-arabinofuranosidase | XP_009612011.1; XP_006351808.1; XP_009762535.1 | Nicotiana tomentosiformis; Solanum tuberosum; Nicotiana sylvestris |
| 4 | HYTAYDIDDWK | beta-xylosidase/alpha-l-arabinofuranosidase | XP_006351808.1 | Solanum tuberosum |
| 5 | YEWWSEALHGISYTGPGVK | beta-xylosidase/alpha-l-arabinofuranosidase | XP_006351808.1; XP_009762535.1 | Solanum tuberosum; Nicotiana sylvestris |
| Glycoside Substrate | Glycosidase Activity (μmol/(h·mg Protein) | Relative Activity (%) |
|---|---|---|
| p-Nitrophenyl β-d-galactopyranoside | 301.35 ± 4.1 | 100 ± 1.6 |
| p-Nitrophenyl β-d-xylopyranoside | 118.62 ± 2.4 | 39.36 ± 0.7 |
| p-Nitrophenyl β-d-mannopyranoside | 0 | 0 |
| p-Nitrophenyl β-d-glucopyranoside | 0 | 0 |
| Substance | Concentration (mM) | Residual Activity (%) |
|---|---|---|
| d-gluconic acid | 10 | 5.7 ± 0.8 |
| 1 | 13.2 ± 0.69 | |
| AgNO3 | 10 | 8.1 ± 0.63 |
| 1 | 11.9 ± 0.6 | |
| HgCl2 | 10 | 6.5 ± 0.97 |
| 1 | 59.6 ± 0.44 | |
| MgCl2 | 10 | 98.9 ± 3.0 |
| 1 | 100.1 ± 3.4 | |
| CuSO4 | 10 | 95.2 ± 1.5 |
| 1 | 100.5 ± 2.0 | |
| MnCl4 | 10 | 103.5 ± 4.2 |
| 1 | 100.6 ± 2.1 | |
| CaCl2 | 10 | 100.5 ± 2.2 |
| 1 | 102.9 ± 1.5 | |
| Control | — | 100 ± 1.0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, B.; Luo, H.; Liu, B.; Li, W.; Ou, S.; Wu, Y.; Zhang, X.; Pang, X.; Zhang, Z. BcXyl, a β-xylosidase Isolated from Brunfelsia Calycina Flowers with Anthocyanin-β-glycosidase Activity. Int. J. Mol. Sci. 2019, 20, 1423. https://doi.org/10.3390/ijms20061423
Dong B, Luo H, Liu B, Li W, Ou S, Wu Y, Zhang X, Pang X, Zhang Z. BcXyl, a β-xylosidase Isolated from Brunfelsia Calycina Flowers with Anthocyanin-β-glycosidase Activity. International Journal of Molecular Sciences. 2019; 20(6):1423. https://doi.org/10.3390/ijms20061423
Chicago/Turabian StyleDong, Boyu, Honghui Luo, Bin Liu, Wenjun Li, Shaojian Ou, Yongyi Wu, Xuelian Zhang, Xuequn Pang, and Zhaoqi Zhang. 2019. "BcXyl, a β-xylosidase Isolated from Brunfelsia Calycina Flowers with Anthocyanin-β-glycosidase Activity" International Journal of Molecular Sciences 20, no. 6: 1423. https://doi.org/10.3390/ijms20061423
APA StyleDong, B., Luo, H., Liu, B., Li, W., Ou, S., Wu, Y., Zhang, X., Pang, X., & Zhang, Z. (2019). BcXyl, a β-xylosidase Isolated from Brunfelsia Calycina Flowers with Anthocyanin-β-glycosidase Activity. International Journal of Molecular Sciences, 20(6), 1423. https://doi.org/10.3390/ijms20061423