The Root Bark of Morus alba L. Suppressed the Migration of Human Non-Small-Cell Lung Cancer Cells through Inhibition of Epithelial–Mesenchymal Transition Mediated by STAT3 and Src
Abstract
:1. Introduction
2. Results
2.1. Identification of Morusin from MEMA through HPLC Analysis
2.2. MEMA Suppressed the Migration of Human NSCLC Cells
2.3. MEMA Suppressed the Invasion of Human NSCLC Cells
2.4. MEMA Suppressed the Activity of STAT3 and Src in Human NSCLC Cells
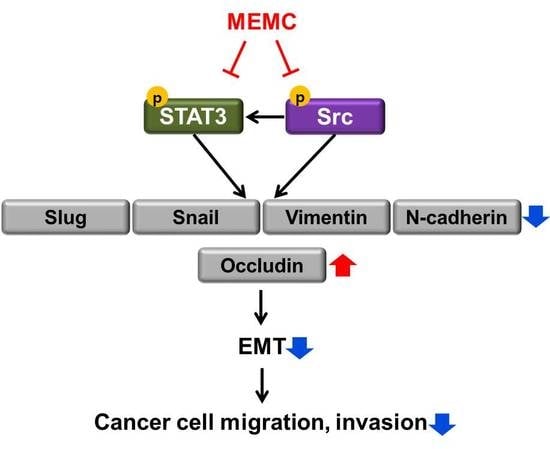
2.5. MEMA Suppressed the Cellular Migration of Human NSCLC Cells through Regulation of EMT Mediated by STAT3 and Src
3. Discussion
4. Materials and Methods
4.1. Preparation of MEMA
4.2. HPLC Analysis
4.3. Cell Culture
4.4. MTT Assay
4.5. Transwell Assay
4.6. Wound-Healing Assay
4.7. Transfection
4.8. Western Blot Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MA | Morus alba L. |
| MEMA | Methylene chloride extract of Morus alba L. |
| EMT | Epithelial–mesenchymal transition |
| HPLC | High-performance liquid chromatography |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (a tetrazole) |
| RT-PCR | Reverse transcriptase-polymerase chain reaction |
| STAT3 | Signal transducer and activator of transcription 3 |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Siegel, R.L.; Jemal, A. Lung Cancer Statistics. Adv. Exp. Med. Biol. 2016, 393, 1–19. [Google Scholar] [CrossRef]
- Perlikos, F.; Harrington, K.J.; Syrigos, K.N. Key molecular mechanisms in lung cancer invasion and metastasis: A comprehensive review. Crit. Rev. Oncol. 2013, 87, 1–11. [Google Scholar] [CrossRef]
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Keith, R.L.; Miller, Y.E. Lung cancer chemoprevention: Current status and future prospects. Nat. Rev. Clin. Oncol. 2013, 10, 334–343. [Google Scholar] [CrossRef]
- Micalizzi, D.S.; Farabaugh, S.M.; Ford, H.L. Epithelial-Mesenchymal Transition in Cancer: Parallels Between Normal Development and Tumor Progression. J. Mammary Gland Biol. Neoplasia 2010, 15, 117–134. [Google Scholar] [CrossRef] [Green Version]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-Mesenchymal Transitions in Development and Disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [Green Version]
- Guadamillas, M.C.; Cerezo, A.; Del Pozo, M.A. Overcoming anoikis—Pathways to anchorage-independent growth in cancer. J. Cell Sci. 2011, 124, 3189–3197. [Google Scholar] [CrossRef]
- Tiwari, N.; Gheldof, A.; Tatari, M.; Christofori, G. EMT as the ultimate survival mechanism of cancer cells. Semin. Cancer Biol. 2012, 22, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Logullo, A.F.; Nonogaki, S.; Pasini, F.S.; Osório, C.A.B.D.T.; Soares, F.A.; Brentani, M.M. Concomitant expression of epithelial-mesenchymal transition biomarkers in breast ductal carcinoma: Association with progression. Oncol. Rep. 2010, 23, 313–320. [Google Scholar] [PubMed]
- Mittal, V. Epithelial Mesenchymal Transition in Aggressive Lung Cancers. Adv. Exp. Med. Biol. 2016, 890, 37–56. [Google Scholar] [CrossRef] [PubMed]
- Kaller, M.; Rokavec, M.; Hörst, D.; Hermeking, H. Pan-cancer EMT-signature identifies RBM47 down-regulation during colorectal cancer progression. Sci. Rep. 2017, 7, 4687. [Google Scholar]
- Batlle, E.; Sancho, E.; Francí, C.; Domínguez, D.; Monfar, M.; Baulida, J.; De Herreros, A.G. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000, 2, 84–89. [Google Scholar] [CrossRef]
- Bolós, V.; Peinado, H.; A Pérez-Moreno, M.; Fraga, M.F.; Esteller, M.; Cano, A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: A comparison with Snail and E47 repressors. J. Cell Sci. 2003, 116, 499–511. [Google Scholar] [CrossRef]
- Vesuna, F.; Van Diest, P.; Chen, J.H.; Raman, V. Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem. Biophys. Commun. 2008, 367, 235–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, K.-O.; Kim, B.-Y.; Lee, M.-H.; Kim, Y.-R.; Chung, H.-Y.; Park, J.-H.; Moon, J.-O. In-vitro and in-vivo anti-inflammatory effect of oxyresveratrol from Morus alba L. J. Pharm. Pharmacol. 2003, 55, 1695–1700. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.-W.; Juang, L.-J.; Wang, B.-S.; Wang, M.-Y.; Tai, H.-M.; Hung, W.-J.; Chen, Y.-J.; Huang, M.-H. Antioxidant and antityrosinase activity of mulberry (Morus alba L.) twigs and root bark. Food Chem. Toxicol. 2011, 49, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chen, M.; Zhang, H.-Q.; Sun, S.; Xia, B.; Wu, F.-H. In vivo hypoglycemic effects of phenolics from the root bark of Morus alba. Fitoterapia 2009, 80, 475–477. [Google Scholar] [CrossRef]
- Nam, S.-Y.; Yi, H.-K.; Lee, J.C.; Kim, J.C.; Song, C.H.; Park, J.W.; Lee, D.Y.; Kim, J.S.; Hwang, P.H. Cortex mori extract induces cancer cell apoptosis through inhibition of microtubule assembly. Arch. Pharmacal Res. 2002, 25, 191–196. [Google Scholar] [CrossRef]
- Yoo, Y.; Park, S.-H.; Chi, G.Y.; Eom, H.S.; Kim, G.-Y.; Hyun, J.W.; Lee, S.-J.; Choi, Y.H. Role of autophagy in apoptosis induction by methylene chloride extracts of Mori cortex in NCI-H460 human lung carcinoma cells. Int. J. Oncol. 2012, 40, 1929–1940. [Google Scholar] [CrossRef]
- Human Metabolome Database. Available online: http://www.hmdb.ca/metabolites/HMDB0036631 (accessed on 28 February 2019).
- Lim, S.L.; Park, S.Y.; Kang, S.; Park, D.; Kim, S.H.; Um, J.Y.; Jang, H.J.; Lee, J.H.; Jeong, C.H.; Jang, J.H.; et al. Morusin induces cell death through inactivating STAT3 signaling in prostate cancer cells. Am. J. Cancer Res. 2015, 5, 289–299. [Google Scholar] [PubMed]
- Kamran, M.Z.; Patil, P.; Gude, R.P. Role of STAT3 in Cancer Metastasis and Translational Advances. BioMed Res. Int. 2013, 2013, 421821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yu, D. Targeting Src family kinases in anti-cancer therapies: Turning promise into triumph. Trends Pharmacol. Sci. 2012, 33, 122–128. [Google Scholar] [CrossRef]
- Huang, S.; Bucana, C.D.; Van Arsdall, M.; Fidler, I.J. Stat1 negatively regulates angiogenesis, tumorigenicity and metastasis of tumor cells. Oncogene 2002, 21, 2504–2512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avalle, L.; Pensa, S.; Regis, G.; Novelli, F.; Poli, V. STAT1 and STAT3 in tumorigenesis: A matter of balance. JAK-STAT 2012, 1, 65–72. [Google Scholar] [CrossRef]
- Wendt, M.K.; Balanis, N.; Carlin, C.R.; Schiemann, W.P. STAT3 and epithelial–mesenchymal transitions in carcinomas. JAK-STAT 2014, 3, e28975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, A.; Sabbineni, H.; Clarke, A.; Somanath, P.R. Novel roles of Src in cancer cell epithelial-to-mesenchymal transition, vascular permeability, microinvasion and metastasis. Life Sci. 2016, 157, 52–61. [Google Scholar] [CrossRef] [Green Version]
- Feng, R.; Liu, X. Inhibition of epithelial to mesenchymal transition in metastatic breast carcinoma cells by c-Src suppression. Acta Biochim. Biophys. Sin. 2010, 42, 496–501. [Google Scholar] [Green Version]
- Li, B.; Huang, C. Regulation of EMT by STAT3 in gastrointestinal cancer (Review). Int. J. Oncol. 2017, 50, 753–767. [Google Scholar] [CrossRef]
- Cheung, K.J.; Ewald, A.J. Illuminating breast cancer invasion: Diverse roles for cell–cell interactions. Cell Biol. 2014, 30, 99–111. [Google Scholar] [CrossRef]
- Schliekelman, M.J.; Taguchi, A.; Zhu, J.; Dai, X.; Rodriguez, J.; Celiktas, M.; Zhang, Q.; Chin, A.; Wong, C.-H.; Wang, H.; et al. Molecular portraits of epithelial, mesenchymal and hybrid states in lung adenocarcinoma and their relevance to survival. Cancer Res. 2015, 75, 1789–1800. [Google Scholar] [CrossRef] [Green Version]
- Jolly, M.K.; Tripathi, S.C.; Jia, D.; Mooney, S.M.; Celiktas, M.; Hanash, S.M.; Mani, S.A.; Pienta, K.J.; Ben-Jacob, E.; Levine, H. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget 2016, 7, 27067–27084. [Google Scholar] [CrossRef] [Green Version]
- Roche, J.; Gemmill, R.M.; Drabkin, H.A.; Mok, S.C. Epigenetic Regulation of the Epithelial to Mesenchymal Transition in Lung Cancer. Mol. Cell. Basis Metastasis 2017, 9, 72. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef]
- Levy, D.E.; Darnell, J.E. STATs: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002, 3, 651–662. [Google Scholar] [CrossRef]
- Schindler, C.; Levy, D.E.; Decker, T. JAK-STAT Signaling: From Interferons to Cytokines. J. Biol. Chem. 2007, 282, 20059–20063. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.-L.; Lai, D.-Y.; Lee, Y.-J.; Chen, N.-F.; Tseng, T.-H. Antitumor progression potential of morusin suppressing STAT3 and NFκB in human hepatoma SK-Hep1 cells. Toxicol. Lett. 2015, 232, 490–498. [Google Scholar] [CrossRef]
- Prasad, S.; Yadav, V.R.; Sung, B.; Reuter, S.; Kannappan, R.; Deorukhkar, A.; Diagaradjane, P.; Wei, C.; Baladandayuthapani, V.; Krishnan, S.; et al. Ursolic Acid Inhibits Growth and Metastasis of Human Colorectal Cancer in an Orthotopic Nude Mouse Model by Targeting Multiple Cell Signaling Pathways: Chemosensitization with Capecitabine. Clin. Cancer 2012, 18, 4942–4953. [Google Scholar] [CrossRef] [Green Version]
- Xiang, L.; Chi, T.; Tang, Q.; Yang, X.; Ou, M.; Chen, X.; Yu, X.; Chen, J.; Ho, R.J.; Shao, J.; et al. A pentacyclic triterpene natural product, ursolic acid and its prodrug US597 inhibit targets within cell adhesion pathway and prevent cancer metastasis. Oncotarget 2015, 6, 9295–9312. [Google Scholar] [CrossRef] [Green Version]






| Inter-Day | Intraday | |||
|---|---|---|---|---|
| RT 1 (min) | RSD 2 (%) | RT 1 (min) | RSD 2 (%) | |
| Morusin | 20.252 | 2.3094 × 10−3 | 20.254 | 0 |
| MEMA | 20.255 | 7.0711 × 10−4 | 20.254 | 1.1547 × 10−3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Min, T.-R.; Park, H.-J.; Park, M.N.; Kim, B.; Park, S.-H. The Root Bark of Morus alba L. Suppressed the Migration of Human Non-Small-Cell Lung Cancer Cells through Inhibition of Epithelial–Mesenchymal Transition Mediated by STAT3 and Src. Int. J. Mol. Sci. 2019, 20, 2244. https://doi.org/10.3390/ijms20092244
Min T-R, Park H-J, Park MN, Kim B, Park S-H. The Root Bark of Morus alba L. Suppressed the Migration of Human Non-Small-Cell Lung Cancer Cells through Inhibition of Epithelial–Mesenchymal Transition Mediated by STAT3 and Src. International Journal of Molecular Sciences. 2019; 20(9):2244. https://doi.org/10.3390/ijms20092244
Chicago/Turabian StyleMin, Tae-Rin, Hyun-Ji Park, Moon Nyeo Park, Bonglee Kim, and Shin-Hyung Park. 2019. "The Root Bark of Morus alba L. Suppressed the Migration of Human Non-Small-Cell Lung Cancer Cells through Inhibition of Epithelial–Mesenchymal Transition Mediated by STAT3 and Src" International Journal of Molecular Sciences 20, no. 9: 2244. https://doi.org/10.3390/ijms20092244
APA StyleMin, T. -R., Park, H. -J., Park, M. N., Kim, B., & Park, S. -H. (2019). The Root Bark of Morus alba L. Suppressed the Migration of Human Non-Small-Cell Lung Cancer Cells through Inhibition of Epithelial–Mesenchymal Transition Mediated by STAT3 and Src. International Journal of Molecular Sciences, 20(9), 2244. https://doi.org/10.3390/ijms20092244