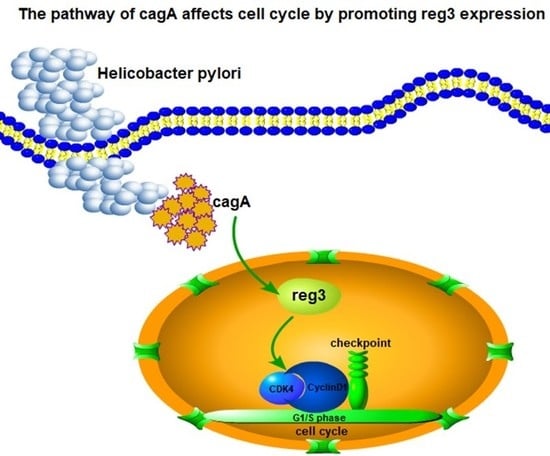
HP-CagA+ Regulates the Expression of CDK4/CyclinD1 via reg3 to Change Cell Cycle and Promote Cell Proliferation
Abstract
:1. Introduction
2. Results
2.1. reg3 Is Highly Expressed in Gastric Cancer and Affects the Prognosis of Patients
2.2. CagA Promotes Cell Proliferation by Regulating reg3
2.3. reg3 Regulates the Growth and Development of Tissues and Organs
2.4. CagA and reg3 Alter the Cell Cycle
2.5. CagA and reg3 Regulate the Expression of CDK4 and CyclinD1
2.6. CagA Regulates the Expression of CDK4 and CyclinD1 through reg3
3. Discussion
4. Materials and Methods
4.1. Experimental Materials
4.2. Hp-CagA Infection in Human Gastric Cancer Cells
4.3. Adeno-Related Carriers, Packaging Cells, and Strains
4.4. Gene Expression Analysis
4.5. Immunohistochemistry
4.6. RNA Extraction and cDNA Synthesis
4.7. Fluorescence qRT-PCR
4.8. Cell Proliferation and Cell Cycle Test
4.9. Immunofluorescence
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ichiyasu, N.; Tada, S.; Kamio, T.; Ueno, N.; Seki, T.; Suko, H. Malignant gastrointestinal stromal tumor of the duodenum after gastrectomy due to early gastric cancer. Report of a case. Jpn. J. Gastro-Enterol. 1998, 95, 1235–1239. [Google Scholar]
- Takahashi, T.; Saikawa, Y.; Igarashi, T.; Tsuwano, S.; Kumagai, K.; Nakamura, R.; Ooyama, T.; Wada, N.; Takeuchi, H.; Takaishi, H.; et al. Octreotide acetate enables the administration of chemoradiotherapy, including the oral anticancer drug S-1, in gastric cancer patients with malignant gastrointestinal obstruction. Oncol. Lett. 2010, 1, 673–677. [Google Scholar]
- Waldum, H.L.; Qvigstad, G.; Sandvik, A.K. Reg protein in gastric cancer tumour cells. FEBS Lett. 2003, 553, 464–465. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Feng, Y.; Hu, Y.; He, C.; Xie, C.; Ouyang, Y.; Artim, S.C.; Huang, D.; Zhu, Y.; Luo, Z.; et al. Helicobacter pylori CagA promotes epithelial mesenchymal transition in gastric carcinogenesis via triggering oncogenic YAP pathway. J. Exp. Clin. Cancer Res. 2018, 37, 280. [Google Scholar] [CrossRef] [Green Version]
- Wakita, A.; Motoyama, S.; Sato, Y.; Koyota, S.; Usami, S.; Yoshino, K.; Sasaki, T.; Imai, K.; Saito, H.; Minamiya, Y. REG Ialpha activates c-Jun through MAPK pathways to enhance the radiosensitivity of squamous esophageal cancer cells. Tumour Biol. 2015, 36, 5249–5254. [Google Scholar] [CrossRef]
- Mitani, Y.; Oue, N.; Matsumura, S.; Yoshida, K.; Noguchi, T.; Ito, M.; Tanaka, S.; Kuniyasu, H.; Kamata, N.; Yasui, W. Reg IV is a serum biomarker for gastric cancer patients and predicts response to 5-fluorouracil-based chemotherapy. Oncogene 2007, 26, 4383–4393. [Google Scholar] [CrossRef] [Green Version]
- Stejskal, D.; Karpisek, M.; Horakova, D.; Cizek, L.; Janoutova, G.; Janout, V. Serum Reg-Ialpha is not suitable marker of metabolic syndrome. Bratisl. Lek. Listy 2007, 108, 138–140. [Google Scholar] [PubMed]
- Cui, W.; Liu, J.L.; Shi, B.Y. Reg family and pancreatic islet beta cell. Sheng Li Ke Xue Jin Zhan 2009, 40, 55–58. [Google Scholar] [PubMed]
- Aida, K.; Saitoh, S.; Nishida, Y.; Yokota, S.; Ohno, S.; Mao, X.; Akiyama, D.; Tanaka, S.; Awata, T.; Shimada, A.; et al. Distinct cell clusters touching islet cells induce islet cell replication in association with over-expression of Regenerating Gene (REG) protein in fulminant type 1 diabetes. PLoS ONE 2014, 9, e95110. [Google Scholar] [CrossRef] [PubMed]
- Bone, A.J.; Banister, S.H.; Zhang, S. The REG gene and islet cell repair and renewal in type 1 diabetes. Adv. Exp. Med. Biol. 1997, 426, 321–327. [Google Scholar]
- Lawniczak, M.; Starzynska, T. Helicobacter pylori CagA(+) infection in gastric cancer patients. Pol. Merkur. Lekarski 2002, 13, 216–220. [Google Scholar]
- Saber, T.; Ghonaim, M.M.; Yousef, A.R.; Khalifa, A.; al Qurashi, H.; Shaqhan, M.; Samaha, M. Association of Helicobacter pylori cagA Gene with Gastric Cancer and Peptic Ulcer in Saudi Patients. J. Microbiol. Biotechnol. 2015, 25, 1146–1153. [Google Scholar] [CrossRef]
- Parsonnet, J.; Friedman, G.D.; Orentreich, N.; Vogelman, H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut 1997, 40, 297–301. [Google Scholar] [CrossRef]
- Sasazuki, S.; Inoue, M.; Iwasaki, M.; Otani, T.; Yamamoto, S.; Ikeda, S.; Hanaoka, T.; Tsugane, S. Japan Public Health Center Study, Effect of Helicobacter pylori infection combined with CagA and pepsinogen status on gastric cancer development among Japanese men and women: A nested case-control study. Cancer Epidemiol. Prev. Biomark. 2006, 15, 1341–1347. [Google Scholar] [CrossRef] [Green Version]
- Fukui, H.; Sekikawa, A.; Chiba, T. REG (regenerating gene) protein--roles of REG protein in H. pylori-induced gastritis and gastric cancers. Nihon Rinsho Jpn. J. Clin. Med. 2005, 11 (Suppl. 63), 98–102. [Google Scholar]
- Kang, D.W.; Hwang, W.C.; Park, M.H.; Ko, G.H.; Ha, W.S.; Kim, K.S.; Lee, Y.C.; Choi, K.Y.; Min, D.S. Rebamipide abolishes Helicobacter pylori CagA-induced phospholipase D1 expression via inhibition of NFkappaB and suppresses invasion of gastric cancer cells. Oncogene 2013, 32, 3531–3542. [Google Scholar] [CrossRef]
- Masu, H. CagA tyrosine phosphorylation motif structure and SHP-2 binding ability of Helicobacter pylori studied in stomach cancer and duodenal ulcer cell lines. Hokkaido J. Med. Sci. 2006, 81, 121–128. [Google Scholar]
- Tsutumi, R.; Hatakeyama, M. Helicobacter pylori CagA and SHP-2 tyrosine phosphatase. Seikagaku 2005, 77, 1269–1273. [Google Scholar]
- Sougleri, I.S.; Papadakos, K.S.; Zadik, M.P.; Mavri-Vavagianni, M.; Mentis, A.F.; Sgouras, D.N. Helicobacter pylori CagA protein induces factors involved in the epithelial to mesenchymal transition (EMT) in infected gastric epithelial cells in an EPIYA- phosphorylation-dependent manner. FEBS J. 2016, 283, 206–220. [Google Scholar] [CrossRef]
- Vaziri, F.; Peerayeh, S.N.; Alebouyeh, M.; Maghsoudi, N.; Azimzadeh, P.; Siadat, S.D.; Zali, M.R. Novel effects of Helicobacter pylori CagA on key genes of gastric cancer signal transduction: A comparative transfection study. Pathog. Dis. 2015, 73, ftu021. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.Y.; Zhang, R.G.; Duan, G.C. Pathogenic mechanisms of the oncoprotein CagA in H. pylori-induced gastric cancer (Review). Oncol. Rep. 2016, 36, 3087–3094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Marhoon, M.S.; Nunn, S.; Soames, R.W. The association between cagA+ H. pylori infection and distal gastric cancer: A proposed model. Dig. Dis. Sci. 2004, 49, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Akhondi-Meybodi, M.; Ghane, M.; Akhondi-Meybodi, S.; Dashti, G. Five-year Survival Rate for Gastric Cancer in Yazd Province, Central Iran, from 2001 to 2008. Middle East J. Dig. Dis. 2017, 9, 39–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, D.M. Assessment of bovine sperm viability by MTT reduction assay. Anim. Reprod. Sci. 2006, 92, 1–8. [Google Scholar] [CrossRef] [PubMed]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Li, X.; Sun, F.; Tong, X.; Bai, Y.; Jin, K.; Liu, L.; Dai, F.; Li, N. HP-CagA+ Regulates the Expression of CDK4/CyclinD1 via reg3 to Change Cell Cycle and Promote Cell Proliferation. Int. J. Mol. Sci. 2020, 21, 224. https://doi.org/10.3390/ijms21010224
Liu B, Li X, Sun F, Tong X, Bai Y, Jin K, Liu L, Dai F, Li N. HP-CagA+ Regulates the Expression of CDK4/CyclinD1 via reg3 to Change Cell Cycle and Promote Cell Proliferation. International Journal of Molecular Sciences. 2020; 21(1):224. https://doi.org/10.3390/ijms21010224
Chicago/Turabian StyleLiu, Bin, Xiaokang Li, Fuze Sun, Xiaoling Tong, Yanmin Bai, Kairang Jin, Lin Liu, Fangyin Dai, and Niannian Li. 2020. "HP-CagA+ Regulates the Expression of CDK4/CyclinD1 via reg3 to Change Cell Cycle and Promote Cell Proliferation" International Journal of Molecular Sciences 21, no. 1: 224. https://doi.org/10.3390/ijms21010224
APA StyleLiu, B., Li, X., Sun, F., Tong, X., Bai, Y., Jin, K., Liu, L., Dai, F., & Li, N. (2020). HP-CagA+ Regulates the Expression of CDK4/CyclinD1 via reg3 to Change Cell Cycle and Promote Cell Proliferation. International Journal of Molecular Sciences, 21(1), 224. https://doi.org/10.3390/ijms21010224