Insights into the FMNAT Active Site of FAD Synthase: Aromaticity Is Essential for Flavin Binding and Catalysis
Abstract
:1. Introduction
2. Results
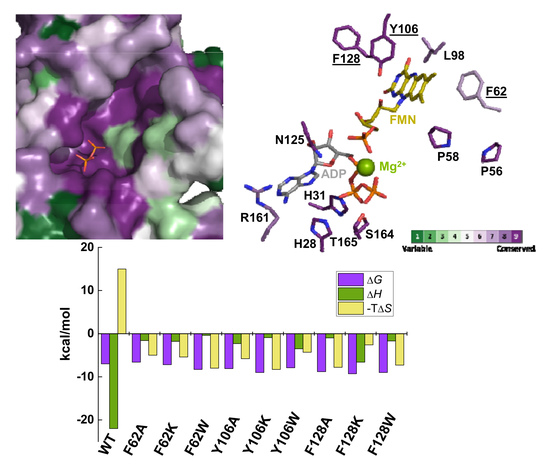
2.1. Substitutions at F62, Y106, and F128 Produce Local Structural Changes
2.2. The Hydrophobic Core at the Isoalloxazine FMNAT site is Essential for FMNAT Activity and Influences the RFK Activity
2.3. The Properties of the FMNAT Isoalloxazine Binding Cavity Determines the Binding Mode of Flavinic Ligands at the FMNAT Module
2.4. The Hydrophobic Nature of F128 Modulates the Binding Mode of the ATP Substrate
3. Discussion
4. Materials and Methods
4.1. Biological Material
4.2. Spectral Analysis
4.3. Qualitative Detection of RFK and FMNAT Activities
4.4. Steady-State Kinetics Parameters for RFK Activities
4.5. Isothermal Titration Calorimetry (ITC)
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ATP | Adenosine 5′-triphosphate |
| CD | Circular dichroism |
| Gdn/HCL | Guanidinium hydrochloride |
| FAD | Flavin adenine dinucleotide, Riboflavin 5′-adenosine diphosphate |
| FADS | FAD synthase |
| FMN | Flavin mononucleotide, Riboflavin 5′-phosphate |
| FMNAT | ATP: FMN adenylyl transferase |
| HPLC | High performance chromatography |
| ITC | Isothermal titration calorimetry |
| PAPS | 3-Phosphoadenosine 5′-phosphosulfate |
| PIPES | 1,4-Piperazinediethanesulfonic acid |
| RF | Riboflavin |
| RFK | ATP: riboflavin kinase |
| TLC | Thin-layer chromatography |
| UV | Ultraviolet |
| WT | Wild-type |
References
- Serrano, A.; Ferreira, P.; Martínez-Júlvez, M.; Medina, M. The prokaryotic FAD synthetase family: A potential drug target. Curr. Pharm. Des. 2013, 19, 2637–2648. [Google Scholar] [CrossRef] [PubMed]
- Sebastián, M.; Anoz-Carbonell, E.; Gracia, B.; Cossio, P.; Aínsa, J.A.; Lans, I.; Medina, M. Discovery of antimicrobial compounds targeting bacterial type FAD synthetases. J. Enzym. Inhib. Med. Chem. 2018, 33, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, S.Y.; Scholle, M.D.; D’Souza, M.; Bernal, A.; Baev, M.V.; Farrell, M.; Kurnasov, O.V.; Daugherty, M.D.; Mseeh, F.; Polanuyer, B.M.; et al. From genetic footprinting to antimicrobial drug targets: Examples in cofactor biosynthetic pathways. J. Bacteriol. 2002, 184, 4555–4572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macheroux, P.; Kappes, B.; Ealick, S.E. Flavogenomics-a genomic and structural view of flavin-dependent proteins. FEBS J. 2011, 278, 2625–2634. [Google Scholar] [CrossRef] [PubMed]
- Joosten, V.; van Berkel, W.J. Flavoenzymes. Curr. Opin. Chem. Biol. 2007, 11, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Huerta, C.; Borek, D.; Machius, M.; Grishin, N.V.; Zhang, H. Structure and mechanism of a eukaryotic FMN adenylyltransferase. J. Mol. Biol. 2009, 389, 388–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frago, S.; Martínez-Júlvez, M.; Serrano, A.; Medina, M. Structural analysis of FAD synthetase from Corynebacterium ammoniagenes. BMC Microbiol. 2008, 8, 160. [Google Scholar] [CrossRef] [Green Version]
- Karthikeyan, S.; Zhou, Q.; Mseeh, F.; Grishin, N.V.; Osterman, A.L.; Zhang, H. Crystal structure of human riboflavin kinase reveals a beta barrel fold and a novel active site arch. Structure 2003, 11, 265–273. [Google Scholar] [CrossRef] [Green Version]
- Herguedas, B.; Lans, I.; Sebastián, M.; Hermoso, J.A.; Martínez-Júlvez, M.; Medina, M. Structural insights into the synthesis of FMN in prokaryotic organisms. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 2526–2542. [Google Scholar] [CrossRef]
- Sebastián, M.; Velázquez-Campoy, A.; Medina, M. The RFK catalytic cycle of the pathogen Streptococcus pneumoniae shows species-specific features in prokaryotic FMN synthesis. J. Enzym. Inhib. Med. Chem. 2018, 33, 842–849. [Google Scholar] [CrossRef] [Green Version]
- Leulliot, N.; Blondeau, K.; Keller, J.; Ulryck, N.; Quevillon-Cheruel, S.; van Tilbeurgh, H. Crystal structure of yeast FAD synthetase (Fad1) in complex with FAD. J. Mol. Biol. 2010, 398, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Lans, I.; Seco, J.; Serrano, A.; Burbano, R.; Cossio, P.; Daza, M.C.; Medina, M. The Dimer-of-Trimers Assembly Prevents Catalysis at the Transferase Site of Prokaryotic FAD Synthase. Biophys. J. 2018, 115, 988–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galluccio, M.; Brizio, C.; Torchetti, E.M.; Ferranti, P.; Gianazza, E.; Indiveri, C.; Barile, M. Over-expression in Escherichia coli, purification and characterization of isoform 2 of human FAD synthetase. Protein Expr. Purif. 2007, 52, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Torchetti, E.M.; Brizio, C.; Colella, M.; Galluccio, M.; Giancaspero, T.A.; Indiveri, C.; Roberti, M.; Barile, M. Mitochondrial localization of human FAD synthetase isoform 1. Mitochondrion 2010, 10, 263–273. [Google Scholar] [CrossRef]
- Torchetti, E.M.; Bonomi, F.; Galluccio, M.; Gianazza, E.; Giancaspero, T.A.; Lametti, S.; Indiveri, C.; Barile, M. Human FAD synthase (isoform 2): A component of the machinery that delivers FAD to apo-flavoproteins. FEBS J. 2011, 278, 4434–4449. [Google Scholar] [CrossRef]
- Giancaspero, T.A.; Galluccio, M.; Miccolis, A.; Leone, P.; Eberini, I.; Iametti, S.; Indiveri, C.; Barile, M. Human FAD synthase is a bi-functional enzyme with a FAD hydrolase activity in the molybdopterin binding domain. Biochem. Biophys. Res. Commun. 2015, 465, 443–449. [Google Scholar] [CrossRef]
- Leone, P.; Galluccio, M.; Barbiroli, A.; Eberini, I.; Tolomeo, M.; Vrenna, F.; Gianazza, E.; Iametti, S.; Bonomi, F.; Indiveri, C.; et al. Bacterial Production, Characterization and Protein Modeling of a Novel Monofuctional Isoform of FAD Synthase in Humans: An Emergency Protein? Molecules 2018, 23, 116. [Google Scholar] [CrossRef] [Green Version]
- Leone, P.; Galluccio, M.; Quarta, S.; Anoz-Carbonell, E.; Medina, M.; Indiveri, C.; Barile, M. Mutation of Aspartate 238 in FAD Synthase Isoform 6 Increases the Specific Activity by Weakening the FAD Binding. Int. J. Mol. Sci. 2019, 20, 6203. [Google Scholar] [CrossRef] [Green Version]
- Miccolis, A.; Galluccio, M.; Nitride, C.; Giancaspero, T.A.; Ferranti, P.; Iametti, S.; Indiveri, C.; Bonomi, F.; Barile, M. Significance of redox-active cysteines in human FAD synthase isoform 2. Biochim. Biophys. Acta 2014, 1844, 2086–2095. [Google Scholar] [CrossRef]
- Miccolis, A.; Galluccio, M.; Giancaspero, T.A.; Indiveri, C.; Barile, M. Bacterial over-expression and purification of the 3′phosphoadenosine 5′phosphosulfate (PAPS) reductase domain of human FAD synthase: Functional characterization and homology modeling. Int. J. Mol. Sci. 2012, 13, 16880–16898. [Google Scholar] [CrossRef] [Green Version]
- Giancaspero, T.A.; Colella, M.; Brizio, C.; Difonzo, G.; Fiorino, G.M.; Leone, P.; Brandsch, R.; Bonomi, F.; Iametti, S.; Barile, M. Remaining challenges in cellular flavin cofactor homeostasis and flavoprotein biogenesis. Front. Chem. 2015, 3, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, Y.; Merrill, A.H., Jr.; McCormick, D.B. Probable reaction mechanisms of flavokinase and FAD synthetase from rat liver. Arch. Biochem. Biophys. 1990, 278, 125–130. [Google Scholar] [CrossRef]
- Sebastián, M.; Lira-Navarrete, E.; Serrano, A.; Marcuello, C.; Velázquez-Campoy, A.; Lostao, A.; Hurtado-Guerrero, R.; Medina, M.; Martínez-Júlvez, M. The FAD synthetase from the human pathogen Streptococcus pneumoniae: A bifunctional enzyme exhibiting activity-dependent redox requirements. Sci. Rep. 2017, 7, 7609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebastián, M.; Arilla-Luna, S.; Bellalou, J.; Yruela, I.; Medina, M. The Biosynthesis of Flavin Cofactors in Listeria monocytogenes. J. Mol. Biol. 2019, 431, 2762–2776. [Google Scholar] [CrossRef]
- Serrano, A.; Sebastián, M.; Arilla-Luna, S.; Baquedano, S.; Herguedas, B.; Velázquez-Campoy, A.; Martínez-Júlvez, M.; Medina, M. The trimer interface in the quaternary structure of the bifunctional prokaryotic FAD synthetase from Corynebacterium ammoniagenes. Sci. Rep. 2017, 7, 404. [Google Scholar] [CrossRef] [Green Version]
- Frago, S.; Velázquez-Campoy, A.; Medina, M. The puzzle of ligand binding to Corynebacterium ammoniagenes FAD synthetase. J. Biol. Chem. 2009, 284, 6610–6619. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Kim, R.; Jancarik, J.; Yokota, H.; Kim, S.H. Crystal structure of a flavin-binding protein from Thermotoga maritima. Proteins 2003, 52, 633–635. [Google Scholar] [CrossRef]
- Herguedas, B.; Martínez-Júlvez, M.; Frago, S.; Medina, M.; Hermoso, J.A. Oligomeric state in the crystal structure of modular FAD synthetase provides insights into its sequential catalysis in prokaryotes. J. Mol. Biol. 2010, 400, 218–230. [Google Scholar] [CrossRef]
- Serrano, A.; Frago, S.; Velázquez-Campoy, A.; Medina, M. Role of key residues at the flavin mononucleotide (FMN):adenylyltransferase catalytic site of the bifunctional riboflavin kinase/flavin adenine dinucleotide (FAD) Synthetase from Corynebacterium ammoniagenes. Int. J. Mol. Sci. 2012, 13, 14492–14517. [Google Scholar] [CrossRef] [Green Version]
- Arilla-Luna, S.; Serrano, A.; Medina, M. Specific Features for the Competent Binding of Substrates at the FMN Adenylyltransferase Site of FAD Synthase from Corynebacterium ammoniagenes. Int. J. Mol. Sci. 2019, 20, 5083. [Google Scholar] [CrossRef] [Green Version]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002; Available online: http://www.pymol.org (accessed on 25 May 2020).
- Serrano, A.; Sebastián, M.; Arilla-Luna, S.; Baquedano, S.; Pallarés, M.C.; Lostao, A.; Herguedas, B.; Velázquez-Campoy, A.; Martínez-Júlvez, M.; Medina, M. Quaternary organization in a bifunctional prokaryotic FAD synthetase: Involvement of an arginine at its adenylyltransferase module on the riboflavin kinase activity. Biochim. Biophys. Acta 2015, 1854, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Sebastián, M.; Serrano, A.; Velázquez-Campoy, A.; Medina, M. Kinetics and thermodynamics of the protein-ligand interactions in the riboflavin kinase activity of the FAD synthetase from Corynebacterium ammoniagenes. Sci. Rep. 2017, 7, 7281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leone, P.; Galluccio, M.; Brizio, C.; Barbiroli, A.; Iametti, S.; Indiveri, C.; Barile, M. The hidden side of the human FAD synthase 2. Int. J. Biol. Macromol. 2019, 138, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Huerta, C.; Grishin, N.V.; Zhang, H. The “super mutant” of yeast FMN adenylyltransferase enhances the enzyme turnover rate by attenuating product inhibition. Biochemistry 2013, 52, 3615–3617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcuello, C.; Arilla-Luna, S.; Medina, M.; Lostao, A. Detection of a quaternary organization into dimer of trimers of Corynebacterium ammoniagenes FAD synthetase at the single-molecule level and at the in cell level. Biochim. Biophys. Acta 2013, 1834, 665–676. [Google Scholar] [CrossRef]
- Gill, S.C.; von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. A Self-Consistent Method for the Analysis of Protein Secondary Structure from Circular Dichroism. Anal. Biochem. 1993, 209, 32–44. [Google Scholar] [CrossRef]
- Johnson, W.C. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins 1999, 25, 307–312. [Google Scholar] [CrossRef]
- Van Stokkum, I.H.; Spoelder, H.J.; Bloemendal, M.; van Grondelle, R.; Groen, F.C. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal. Biochem. 1990, 191, 110–118. [Google Scholar] [CrossRef]
- Leskovac, V. Comprehensive Enzyme Kinetics; Kluwer Adacemic/Plenum Publishers: New York, NY, USA, 2003. [Google Scholar]









| CaFADS | kcat (min−1) | KmFMN (µM) | KmATP (µM) | kcat/KmFMN (min−1·µM−1) | kcat/KmATP (min−1·µM−1) |
|---|---|---|---|---|---|
| WT | 39 ± 1 | 6 ± 1 | 43 ± 8 | 6 ± 1 | 0.9 ± 0.2 |
| F62W | 29 ± 1 | 42 ± 2 | 51 ± 7 | 0.7 ± 0.1 | 0.6 ± 0.1 |
| Y106W | 36 ± 1 | 12 ± 1 | 19 ± 2 | 3.6 ± 0.5 | 2.3 ± 0.2 |
| F128W | 26 ± 1 | 108 ± 13 | 9 ± 2 | 0.2 ± 0.1 | 2.9 ± 0.7 |
| CaFADS | kcatapp (min−1) | KmRF (µM) | Ki (µM) | kcatapp/KmRF (min−1·µM−1) | kcatapp (min−1) | KmATP (µM) | kcatapp/mATP (min−1·µM−1) |
|---|---|---|---|---|---|---|---|
| WT a | 408 ± 230 | 12 ± 3 | 4.9 ± 3.9 | 35 ± 22 | 155 ± 5 | 28 ± 4 | 5.5 ± 0.8 |
| F62A | 284 ± 31 | 2.5 ± 0.7 | 67 ± 22 | 1115 ± 33 | 127 ± 1 | 14 ± 1 | 9.3 ± 0.6 |
| F62K | 131 ± 3 | 0.2 ± 0.1 | 451 ± 131 | 534 ± 70 | 138 ± 5 | 18 ± 3 | 7.6 ± 1.2 |
| F62W | 151 ± 36 | 4.4 ± 1.8 | 20 ± 9 | 34 ± 16 | 92 ± 2 | 18 ± 2 | 5.2 ± 0.5 |
| Y106A | 143 ± 20 | 2.5 ± 0.8 | 34 ± 11 | 58 ± 19 | 53 ± 2 | 29 ± 4 | 1.8 ± 0.3 |
| Y106K | 148 ± 23 | 2.1 ± 0.7 | 23 ± 7 | 71 ± 26 | 54 ± 2 | 39 ± 6 | 1.4 ± 0.2 |
| Y106W | 42 ± 7 | 0.6 ± 0.3 | 34 ± 16 | 68 ± 37 | 21 ± 1 | 49 ± 7 | 0.4 ± 0.1 |
| F128A | 105 ± 20 | 2.6 ± 1.1 | 37 ± 18 | 40 ± 19 | 35 ± 1 | 30 ± 5 | 1.2 ± 0.2 |
| F128K | 68 ± 7 | 1.7 ± 0.5 | 98 ± 43 | 40 ± 12 | 51 ± 2 | 30 ± 4 | 1.7 ± 0.3 |
| F128W | 125 ± 24 | 4.3 ± 1.4 | 21 ± 8 | 29 ± 11 | 70 ± 2 | 38 ± 4 | 1.9 ± 0.2 |
| KdFMN (μM) | KdFAD (μM) | KdATP (μM) | |
|---|---|---|---|
| MgCl2 | 10 mM | 10 mM | — |
| WT | 7.1 ± 0.4 | 1.2 ± 0.1 | 48 ± 7 |
| F62A | 15 ± 1 | 7.9 ± 0.7 | 44 ± 5 |
| F62K | 5.1 ± 0.3 | 75 ± 3 | 54 ± 9 |
| F62W | 0.8 ± 0.1 | 5.4 ± 0.1 | 74 ± 13 |
| Y106A | 1.2 ± 0.1 | 2.6 ± 0.1 | 31 ± 3 |
| Y106K | 0.3 ± 0.1 | 1.3 ± 0.1 | 58 ± 13 |
| Y106W | 1.7 ± 0.1 | 6.2 ± 0.2 | 44 ± 4 |
| F128A | 0.4 ± 0.1 | 14 ± 1 | 33 ± 3 |
| F128K | 0.2 ± 0.1 | 2.0 ± 0.1 | 14 ± 1 |
| F128W | 0.3 ± 0.1 | 2.5 ± 0.1 | 19 ± 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serrano, A.; Arilla-Luna, S.; Medina, M. Insights into the FMNAT Active Site of FAD Synthase: Aromaticity Is Essential for Flavin Binding and Catalysis. Int. J. Mol. Sci. 2020, 21, 3738. https://doi.org/10.3390/ijms21103738
Serrano A, Arilla-Luna S, Medina M. Insights into the FMNAT Active Site of FAD Synthase: Aromaticity Is Essential for Flavin Binding and Catalysis. International Journal of Molecular Sciences. 2020; 21(10):3738. https://doi.org/10.3390/ijms21103738
Chicago/Turabian StyleSerrano, Ana, Sonia Arilla-Luna, and Milagros Medina. 2020. "Insights into the FMNAT Active Site of FAD Synthase: Aromaticity Is Essential for Flavin Binding and Catalysis" International Journal of Molecular Sciences 21, no. 10: 3738. https://doi.org/10.3390/ijms21103738
APA StyleSerrano, A., Arilla-Luna, S., & Medina, M. (2020). Insights into the FMNAT Active Site of FAD Synthase: Aromaticity Is Essential for Flavin Binding and Catalysis. International Journal of Molecular Sciences, 21(10), 3738. https://doi.org/10.3390/ijms21103738