Small Nucleolar RNAs Determine Resistance to Doxorubicin in Human Osteosarcoma
Abstract
:1. Introduction
2. Results
2.1. snoRNA Family is Up-Regulated in Doxorubicin-Resistant Osteosarcoma Cells
2.2. SNORD3A, SNORA13 and SNORA28 Are Ontologically and Functionally Related to Doxorubicin Resistance in Osteosarcoma Cells
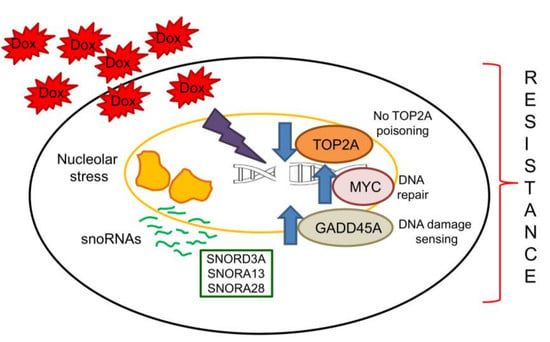
2.3. SNORD3A, SNORA13 and SNORA28 Contribute to Doxorubicin Resistance by Up-Regulating GADD45A and c-MYC, and Down-Regulating Topoisomerase 2A
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cells
4.3. Gene Expression Profiling Analysis
4.4. SnoRNA Overexpression
4.5. Cell Silencing/Overexpression
4.6. RT-PCR and PCR Array
4.7. Doxorubicin Intracellular Accumulation
4.8. Immunoblot
4.9. LDH Release
4.10. Cell Viability
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Dox | doxorubicin |
| ABCB1 | ATP binding cassette transporter B1 |
| Pgp | P-glycoprotein |
| lncRNA | long non-coding RNA |
| miRNA | microRNA |
| LUCAT1 | lung cancer associated transcript 1 |
| LINC00161 | long intergenic non-protein coding RNA 161 |
| IFIT-2 | interferon-induced protein with tetratricopeptide repeats 2 |
| SNHG | small nucleolar RNA host gene |
| MCL1 | myeloid cell leukemia 1 |
| ATG4B | autophagy-related 4B |
| snoRNA | small nucleolar RNA |
| SNORA | box H/ACA snoRNA |
| rRNA | ribosomal RNA |
| SNORD | box C/D snoRNA |
| mTOR | mammalian target of rapamycin |
| p70 S6K | p70 S6 kinase |
| eIF4E-BP1 | eukaryotic initiation factor 4E-binding protein 1 |
| Rp113a | ribosomal protein L13a |
| RRP12 | ribosomal RNA processing 12 homolog |
| RACK1 | receptor for activated C-kinase 1 |
| OSCP1 | organic solute carrier partner 1 |
| EPB41L4A | erythrocyte membrane protein band 4.1 like 4A |
| EIF5 | eukaryotic translation initiation factor-5 |
| LDH | lactate dehydrogenase |
| TOP2A | topoisomerase 2A |
| 5-FU | 5-fluorouracil |
| ERRα | estrogen-related receptor α |
| NRF2 | nuclear factor (erythroid-derived 2)-like 2 |
References
- Yang, J.; Zhang, W. New molecular insights into osteosarcoma targeted therapy. Curr. Opin. Oncol. 2013, 25, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Hattinger, C.M.; Fanelli, M.; Tavanti, E.; Vella, S.; Riganti, C.; Picci, P.; Serra, M. Doxorubicin-resistant osteosarcoma: Novel therapeutic approaches in sight? Future Oncol. 2017, 13, 673–677. [Google Scholar] [CrossRef] [PubMed]
- Hattinger, C.M.; Patrizio, M.P.; Magagnoli, F.; Luppi, S.; Serra, M. An update on emerging drugs in osteosarcoma: Towards tailored therapies? Expert Opin. Emerg. Drugs. 2019, 24, 153–171. [Google Scholar] [CrossRef]
- He, H.; Ni, J.; Huang, J. Molecular mechanisms of chemoresistance in osteosarcoma. Oncol. Lett. 2014, 7, 1352–1362. [Google Scholar] [CrossRef] [Green Version]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldini, N.; Scotlandi, K.; Barbanti-Bròdano, G.; Manara, M.C.; Maurici, D.; Bacci, G.; Bertoni, F.; Picci, P.; Sottili, S.; Camapnacci, M.; et al. Expression of P-glycoprotein in high-grade osteosarcomas in relation to clinical outcome. N. Engl. J. Med. 1995, 333, 1380–1385. [Google Scholar] [CrossRef] [PubMed]
- Serra, M.; Scotlandi, K.; Reverter-Branchat, G.; Ferrari, S.; Manara, M.C.; Benini, S.; Incaprera, M.; Bertoni, F.; Mercuri, M.; Briccoli, A.; et al. Value of P-glycoprotein and clinicopathologic factors as the basis for new treatment strategies in high-grade osteosarcoma of the extremities. J. Clin. Oncol. 2003, 21, 536–542. [Google Scholar] [CrossRef]
- Serra, M.; Pasello, M.; Manara, M.C.; Scotlandi, K.; Ferrari, S.; Bertoni, F.; Mercuri, M.; Alvegard, T.A.; Picci, P.; Bacci, G.; et al. May P-glycoprotein status be used to stratify high-grade osteosarcoma patients? Results from the Italian/Scandinavian Sarcoma Group 1 treatment protocol. Int. J. Oncol. 2006, 29, 1459–1468. [Google Scholar] [CrossRef]
- Hattinger, C.M.; Patrizio, M.P.; Tavanti, E.; Luppi, S.; Magagnoli, F.; Picci, P.; Serra, M. Genetic testing for high-grade osteosarcoma: A guide for future tailored treatments? Expert Rev. Mol. Diagn. 2018, 18, 947–961. [Google Scholar] [CrossRef]
- Wang, J.Y.; Yang, Y.; Ma, Y.; Wang, F.; Xue, A.; Zhu, J.; Yang, H.; Chen, Q.; Chen, M.; Ye, L.; et al. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed. Pharmacother. 2020, 121, 109627. [Google Scholar] [CrossRef]
- Han, Z.; Shi, L. Long non-coding RNA LUCAT1 modulates methotrexate resistance in osteosarcoma via miR-200c/ABCB1 axis. Biochem. Biophys. Res. Commun. 2018, 495, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, L.; Zheng, X.; Zhong, W.; Tian, X.; Yin, B.; Tian, K.; Zhang, W. Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett. 2016, 382, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Li, L.; Li, Y.; Sun, H.; Zeng, C. Long noncoding RNA SNHG12 mediates doxorubicin resistance of osteosarcoma via miR-320a/MCL1 axis. Biomed. Pharmacother. 2018, 106, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gu, S.; Li, H.; Wang, J.; Wei, C.; Liu, Q. SNHG16 promotes osteosarcoma progression and enhances cisplatin resistance by sponging miR-16 to upregulate ATG4B expression. Biochem. Biophys. Res. Commun. 2019, 518, 127–133. [Google Scholar] [CrossRef]
- Bergeron, D.; Fafard-Couture, É.; Scott, M.S. Small nucleolar RNAs: Continuing identification of novel members and increasing diversity of their molecular mechanisms of action. Biochem. Soc. Trans. 2020, 48, 645–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, Y.H.; Chen, L.L. Processing and roles of snoRNA ended long noncoding RNA. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 596–606. [Google Scholar] [CrossRef]
- Kiss, T.; Fayet, E.; Jády, B.E.; Richard, P.; Weber, M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 407–417. [Google Scholar] [CrossRef] [Green Version]
- Bratkovič, T.; Božič, J.; Rogelj, B. Functional diversity of small nucleolar RNAs. Nucleic Acids Res. 2020, 48, 1627–1651. [Google Scholar] [CrossRef] [Green Version]
- Brameier, M.; Herwig, A.; Reinhardt, R.; Walter, L.; Gruber, J. Human box C/D snoRNAs with miRNA like functions: Expanding the range of regulatory RNAs. Nucleic Acids Res. 2011, 39, 675–686. [Google Scholar] [CrossRef]
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small non-coding RNA and cancer. Carcinogenesis 2017, 38, 485–491. [Google Scholar] [CrossRef] [Green Version]
- Deschamps-Francoeur, G.; Garneau, D.; Dupuis-Sandoval, F.; Roy, A.; Frappier, M.; Catala, M.; Couture, S.; Barbe-Marcoux, M.; Abou-Elela, S.; Scott, M.S. Identification of discrete classes of small nucleolar RNA featuring different ends and RNA binding protein dependency. Nucleic Acids Res. 2014, 42, 10073–10085. [Google Scholar] [CrossRef] [PubMed]
- Kufel, J.; Grzechnik, P. Small Nucleolar RNAs Tell a Different Tale. Trends Genet. 2019, 35, 104–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abel, Y.; Rederstorff, M. SnoRNAs and the emerging class of sdRNAs: Multifaceted players in oncogenesis. Biochimie 2019, 164, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Deogharia, M.; Majumder, M. Guide snoRNAs: Drivers or Passengers in Human Disease? Biology 2018, 8, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.P.; Zhang, B.X.; Chu, L. Small Nucleolar RNAs: Insight Into Their Function in Cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, L.; Yi, H.; Chen, C.; Yan, S.; Yao, H.; He, G.; Li, G.; Jiang, Y.; Deng, T.; Deng, X. Nucleolar stress: Is there a reverse version? J. Cancer 2018, 9, 3723–3727. [Google Scholar] [CrossRef]
- Taymaz-Nikerel, H.; Karabekmez, M.E.; Eraslan, S.; Kırdar, B. Doxorubicin induces an extensive transcriptional and metabolic rewiring in yeast cells. Sci. Rep. 2018, 8, 13672. [Google Scholar] [CrossRef]
- Halim, V.A.; García-Santisteban, I.; Warmerdam, D.O.; van den Broek, B.; Heck, A.J.R.; Mohammed, S.; Medema, R.H. Doxorubicin-induced DNA Damage Causes Extensive Ubiquitination of Ribosomal Proteins Associated with a Decrease in Protein Translation. Mol. Cell. Proteom. 2018, 17, 2297–2308. [Google Scholar] [CrossRef] [Green Version]
- Gaur, S.; Chen, L.; Yang, L.; Wu, X.; Un, F.; Yen, Y. Inhibitors of mTOR overcome drug resistance from topoisomerase II inhibitors in solid tumors. Cancer Lett. 2011, 311, 20–28. [Google Scholar] [CrossRef]
- Holley, C.L.; Li, M.W.; Scruggs, B.S.; Matkovich, S.J.; Ory, D.S.; Schaffer, J.E. Cytosolic accumulation of small nucleolar RNAs (snoRNAs) is dynamically regulated by NADPH oxidase. J. Biol. Chem. 2015, 290, 11741–11748. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.J.; Lee, H.W.; Lee, Y.S.; Shim, D.M.; Seo, S.W. RRP12 is a crucial nucleolar protein that regulates p53 activity in osteosarcoma cells. Tumour Biol. 2016, 37, 4351–4358. [Google Scholar] [CrossRef]
- Ruan, Y.; Sun, L.; Hao, Y.; Wang, L.; Xu, J.; Zhang, W.; Xie, J.; Guo, L.; Zhou, L.; Yun, X.; et al. Ribosomal RACK1 promotes chemoresistance and growth in human hepatocellular carcinoma. J. Clin. Investig. 2012, 122, 2554–2566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serra, M.; Scotlandi, K.; Manara, M.C.; Maurici, D.; Lollini, P.L.; De Giovanni, C.; Toffoli, G.; Baldini, N. Establishment and characterization of multidrug-resistant human osteosarcoma cell lines. Anticancer Res. 1993, 13, 323–329. [Google Scholar] [PubMed]
- Lestrade, L.; Weber, M.J. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006, 34, D158–D162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvador, J.M.; Brown-Clay, J.D.; Fornace, A.J., Jr. Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv. Exp. Med. Biol. 2013, 793, 1–19. [Google Scholar] [CrossRef]
- Chen, B.J.; Wu, Y.L.; Tanaka, Y.; Zhang, W. Small molecules targeting c-Myc oncogene: Promising anti-cancer therapeutics. Int. J. Biol. Sci. 2014, 10, 1084–1096. [Google Scholar] [CrossRef]
- Roca, J. Topoisomerase II: A fitted mechanism for the chromatin landscape. Nucleic Acids Res. 2009, 37, 721–730. [Google Scholar] [CrossRef] [Green Version]
- Pakos, E.E.; Ioannidis, J.P. The association of P-glycoprotein with response to chemotherapy and clinical outcome in patients with osteosarcoma. A meta-analysis. Cancer 2003, 98, 581–589. [Google Scholar] [CrossRef]
- Buondonno, I.; Gazzano, E.; Tavanti, E.; Chegaev, K.; Kopecka, J.; Fanelli, M.; Rolando, B.; Fruttero, R.; Gasco, A.; Hattinger, C.; et al. Endoplasmic reticulum-targeting doxorubicin: A new tool effective against doxorubicin-resistant osteosarcoma. Cell. Mol. Life Sci. 2019, 76, 609–625. [Google Scholar] [CrossRef]
- Huu, N.T.; Yoshida, H.; Yamaguchi, M. Tumor suppressor gene OSCP1/NOR1 regulates apoptosis, proliferation, differentiation, and ROS generation during eye development of Drosophila melanogaster. FEBS J. 2015, 282, 4727–4746. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, Y.; Shibusawa, A.; Saito, H.; Ohshiro, N.; Ohbayashi, M.; Kohyama, N.; Yamamoto, T. Isolation and functional characterization of a novel organic solute carrier protein, hOSCP1. J. Biol. Chem. 2005, 280, 32332–32339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishiguro, H.; Furukawa, Y.; Daigo, Y.; Miyoshi, Y.; Nagasawa, Y.; Nishiwaki, T.; Kawasoe, T.; Fujita, M.; Satoh, S.; Miwa, N.; et al. Isolation and characterization of human NBL4, a gene involved in the beta-catenin/tcf signaling pathway. Jpn. J. Cancer Res. 2000, 91, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Dever, T.E.; Dinman, J.D.; Green, R. Translation Elongation and Recoding in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10, a032649. [Google Scholar] [CrossRef] [PubMed]
- Roychowdhury, A.; Samadder, S.; Das, P.; Mazumder, D.I.; Chatterjee, A.; Addya, S.; Mondal, R.; Roy, A.; Roychoudhury, S.; Panda, C.K. Deregulation of H19 is associated with cervical carcinoma. Genomics 2020, 112, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhang, J.; Tang, H.; Zhai, D.; Huang, D.; Ling, L.; Wang, X.; Liu, T.; Zhang, Q.; Zhang, Z.; et al. LncRNA SNORD3A specifically sensitizes breast cancer cells to 5-FU by sponging miR-185-5p to enhance UMPS expression. Cell Death Dis. 2020, 11, 329. [Google Scholar] [CrossRef] [PubMed]
- Montag, G.; Bankoglu, E.E.; Bolte, A.; Hintzsche, H.; Djelic, N.; Stopper, H. Differentiated and exponentially growing HL60 cells exhibit different sensitivity to some genotoxic agents in the comet assay. Mutat. Res. 2019, 845, 402972. [Google Scholar] [CrossRef]
- Gazitt, Y.; Kolaparthi, V.; Moncada, K.; Thomas, C.; Freeman, J. Targeted therapy of human osteosarcoma with 17AAG or rapamycin: Characterization of induced apoptosis and inhibition of mTOR and Akt/MAPK/Wnt pathways. Int. J. Oncol. 2009, 34, 551–561. [Google Scholar] [CrossRef] [Green Version]
- Guerzoni, C.; Amatori, S.; Giorgi, L.; Manara, M.C.; Landuzzi, L.; Lollini, P.L.; Tassoni, A.; Balducci, M.; Manfrini, M.; Pratelli, L.; et al. An aza-macrocycle containing maltolic side-arms (maltonis) as potential drug against human pediatric sarcomas. BMC Cancer 2014, 14, 137. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.R.; Xiong, Y.; Duan, H.; Gong, R.R. Identification of genes associated with methotrexate resistance in methotrexate-resistant osteosarcoma cell lines. J. Orthop. Surg. Res. 2015, 10, 136. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, N.; Nobusue, H.; Shimizu, T.; Sugihara, E.; Yamaguchi-Iwai, S.; Onishi, N.; Kunitomi, H.; Kuroda, T.; Saya, H. ROCK Inhibition Induces Terminal Adipocyte Differentiation and Suppresses Tumorigenesis in Chemoresistant Osteosarcoma Cells. Cancer Res. 2019, 79, 3088–3099. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Maacha, S.; Belkhiri, A. Transcriptional upregulation of c-MYC by AXL confers epirubicin resistance in esophageal adenocarcinoma. Mol. Oncol. 2018, 12, 2191–2208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.C.; Shi, L.H.; Wang, X.J.; Wang, S.X.; Wan, X.Q.; Liu, S.R.; Wang, Y.F.; Lu, Z.; Wang, L.H.; Ding, Y. Stat3/Oct-4/c-Myc signal circuit for regulating stemness-mediated doxorubicin resistance of triple-negative breast cancer cells and inhibitory effects of WP1066. Int. J. Oncol. 2018, 53, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Folk, W.P.; Sakamuro, D. The Dual Roles of MYC in Genomic Instability and Cancer Chemoresistance. Genes 2017, 8, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morcelle, C.; Menoyo, S.; Morón-Duran, F.D.; Tauler, A.; Kozma, S.C.; Thomas, G.; Gentilella, A. Oncogenic MYC Induces the Impaired Ribosome Biogenesis Checkpoint and Stabilizes p53 Independent of Increased Ribosome Content. Cancer Res. 2019, 79, 4348–4359. [Google Scholar] [CrossRef] [Green Version]
- Erriquez, J.; Becco, P.; Olivero, M.; Ponzone, R.; Maggiorotto, F.; Ferrero, A.; Scalzo, M.S.; Canuto, E.M.; Sapino, A.; Verdun di Cantogno, L.; et al. TOP2A gene copy gain predicts response of epithelial ovarian cancers to pegylated liposomal doxorubicin: TOP2A as marker of response to PLD in ovarian cancer. Gynecol. Oncol. 2015, 138, 627–633. [Google Scholar] [CrossRef]
- Zhao, M.; Yu, S.; Zhang, M. Differential expression of multidrug resistance-related proteins in adriamycin-resistant (pumc-91/ADM) and parental (pumc-91) human bladder cancer cell lines. Mol. Med. Rep. 2016, 14, 4741–4746. [Google Scholar] [CrossRef]
- Jeon, K.H.; Yu, H.V.; Kwon, Y. Hyperactivated m-calpain affects acquisition of doxorubicin resistance in breast cancer cells. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1126–1133. [Google Scholar] [CrossRef]
- Kumar, A.; Ehrenshaft, M.; Tokar, E.J.; Mason, R.P.; Sinha, B.K. Nitric oxide inhibits topoisomerase II activity and induces resistance to topoisomerase II-poisons in human tumor cells. Biochim. Biophys. Acta 2016, 1860, 1519–1527. [Google Scholar] [CrossRef] [Green Version]
- Susa, M.; Iyer, A.K.; Ryu, K.; Choy, E.; Hornicek, F.J.; Mankin, H.; Milane, L.; Amiji, M.M.; Duan, Z. Inhibition of ABCB1 (MDR1) expression by an siRNA nanoparticulate delivery system to overcome drug resistance in osteosarcoma. PLoS ONE 2010, 5, e10764. [Google Scholar] [CrossRef] [Green Version]
- Perez, J.; Bardin, C.; Rigal, C.; Anthony, B.; Rousseau, R.; Dutour, A. Anti-MDR1 siRNA restores chemosensitivity in chemoresistant breast carcinoma and osteosarcoma cell lines. Anticancer Res. 2011, 31, 2813–2820. [Google Scholar]
- Chen, Y.; Zhang, K.; Li, Y.; Guo, R.; Zhang, K.; Zhong, G.; He, Q. Oestrogen-related receptor alpha mediates chemotherapy resistance of osteosarcoma cells via regulation of ABCB1. J. Cell. Mol. Med. 2019, 23, 2115–2124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.C.; Tu, M.J.; Ho, P.Y.; Jilek, J.L.; Duan, Z.; Zhang, Q.Y.; Yu, A.X.; Yu, A.M. Bioengineered NRF2-siRNA Is Effective to Interfere with NRF2 Pathways and Improve Chemosensitivity of Human Cancer Cells. Drug Metab. Dispos. 2018, 46, 2–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senapati, D.; Patra, B.C.; Kar, A.; Chini, D.S.; Ghosh, S.; Patra, S.; Bhattacharya, M. Promising approaches of small interfering RNAs (siRNAs) mediated cancer gene therapy. Gene 2019, 719, 144071. [Google Scholar] [CrossRef]
- Jin, J.O.; Kim, G.; Hwang, J.; Han, K.H.; Kwak, M.; Lee, P.C.W. Nucleic acid nanotechnology for cancer treatment. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188377. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Riganti, C.; Salaroglio, I.C.; Caldera, V.; Campia, I.; Kopecka, J.; Mellai, M.; Annovazzi, L.; Bosia, A.; Ghigo, D.; Schiffer, D. Temozolomide downregulates P-glycoprotein expression in glioblastoma stem cells by interfering with the Wnt3a/glycogen synthase-3 kinase/β-catenin pathway. Neuro Oncol. 2013, 15, 1502–1517. [Google Scholar] [CrossRef] [Green Version]






| Gene | Primers |
|---|---|
| SNORD3A_BAMHI_F | GCGGATCCAAGACTATACTTTCAGGGATCATTTC |
| SNORD3A_XHOI_R | CGCTCGAGACCACTCAGACCGCGTT |
| SNORA13_BAMHI_F | GCGGATCCAGCCTTTGTGTTGCC |
| SNORA13_XHOI_R | CGCTCGAGAACTGTTACTTATGCAGCTCC |
| SNORA28_BAMHI_F | GCGGATCCAAGCAACACTCTGTG |
| SNORA28_XHOI_R | CGCTCGAGACTGTTTAAGTCTATATAACGGC |
| Gene | Primers |
|---|---|
| SNORD3A_F | TAGAGCACCGAAAACCACGA |
| SNORD3A_R | CCTCTCACTCCCCAATACGG |
| OSCP1_F | ATCAACATACAAGCCACCCAG |
| OSCP1_R | ATCATGGCGAGCAAATCGTC |
| SNORA13_F | TTTGTGTTGCCCATTCACTTTG |
| SNORA13_R | ACTTATGCAGCTCCTACACCAA |
| EPB41L4A_F | AAGCAGCGTGCCTGGTTAC |
| EPB41L4A_R | ATGCTCCCAGATGGTATTCAGC |
| SNORA28_F | GTCTGACACAATTTGAGCTTGCT |
| SNORA28_R | ATAACGGCTTGTCTCATGGGA |
| EIF5_F | CACCTGAGAATAGTGACAGTGGT |
| EIF5_R | TCATTTGGTGGTGGTGGTGG |
| GADD45A_F | TGCTCAGCAAAGCCCTGAGT |
| GADD45A_R | GCAGGCACAACACCACGTTA |
| MYC_F | ACCACCAGCAGCGACTCTGA |
| MYC_R | TCCAGCAGAAGGTGATCCAGACT |
| TOP2A_F | CTAGTTAATGCTGCGGACAACA |
| TOP2A_R | CATTTCGACCACCTGTCACTT |
| S14_F | CGAGGCTGATGACCTGTTCT |
| S14_R | GCCCTCTCCCACTCTCTCTT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Godel, M.; Morena, D.; Ananthanarayanan, P.; Buondonno, I.; Ferrero, G.; Hattinger, C.M.; Di Nicolantonio, F.; Serra, M.; Taulli, R.; Cordero, F.; et al. Small Nucleolar RNAs Determine Resistance to Doxorubicin in Human Osteosarcoma. Int. J. Mol. Sci. 2020, 21, 4500. https://doi.org/10.3390/ijms21124500
Godel M, Morena D, Ananthanarayanan P, Buondonno I, Ferrero G, Hattinger CM, Di Nicolantonio F, Serra M, Taulli R, Cordero F, et al. Small Nucleolar RNAs Determine Resistance to Doxorubicin in Human Osteosarcoma. International Journal of Molecular Sciences. 2020; 21(12):4500. https://doi.org/10.3390/ijms21124500
Chicago/Turabian StyleGodel, Martina, Deborah Morena, Preeta Ananthanarayanan, Ilaria Buondonno, Giulio Ferrero, Claudia M. Hattinger, Federica Di Nicolantonio, Massimo Serra, Riccardo Taulli, Francesca Cordero, and et al. 2020. "Small Nucleolar RNAs Determine Resistance to Doxorubicin in Human Osteosarcoma" International Journal of Molecular Sciences 21, no. 12: 4500. https://doi.org/10.3390/ijms21124500
APA StyleGodel, M., Morena, D., Ananthanarayanan, P., Buondonno, I., Ferrero, G., Hattinger, C. M., Di Nicolantonio, F., Serra, M., Taulli, R., Cordero, F., Riganti, C., & Kopecka, J. (2020). Small Nucleolar RNAs Determine Resistance to Doxorubicin in Human Osteosarcoma. International Journal of Molecular Sciences, 21(12), 4500. https://doi.org/10.3390/ijms21124500