Differential Profiles of Gut Microbiota and Metabolites Associated with Host Shift of Plutella xylostella
Abstract
:1. Introduction
2. Results
2.1. Diversity of Gut Microbiota in P. xylostella Populations from Different Hosts
2.2. Abundance of Each Bacterial Family in the Gut Microbiota of P. xylostella Populations from Different Hosts
2.3. Functional Enrichment Profiles of Gut Microbes
2.4. Profiling of the Midgut Metabolites in PxR and PxP-17
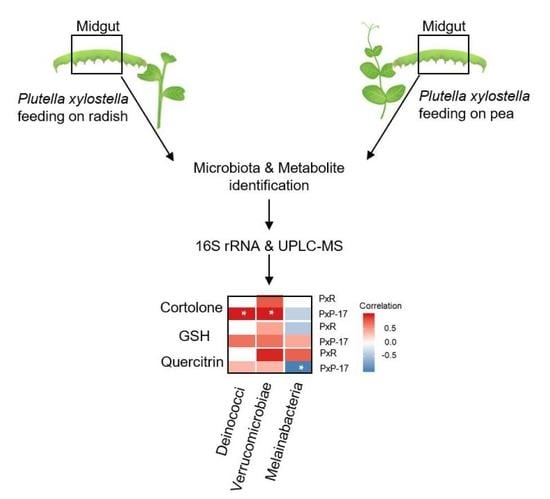
2.5. Associations between Gut Microbes and Metabolites in P. xylostella
3. Discussion
4. Materials and Methods
4.1. Insect Rearing
4.2. Sample Collection
4.3. 16S rDNA Sequencing
4.4. Metabolite Extraction and Profiling
4.5. Data Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- KesÏnerova, L.; Mars, R.A.T.; Ellegaard, K.M.; Kes, L.; Sauer, U.; Engel, P. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 2017, 15, e2003467. [Google Scholar]
- Schneider, H.; Simmering, R.; Hartmann, L.; Pforte, H.; Blaut, M. Degradation of quercetin-3-glucoside in gnotobiotic rats associated with human intestinal bacteria. J. Appl. Microbiol. 2000, 89, 1027–1037. [Google Scholar] [CrossRef]
- Braune, A.; Gütschow, M.; Engst, W.; Blaut, M. Degradation of quercetin and luteolin by Eubacterium ramulus. Appl. Environ. Microbiol. 2001, 67, 5558–5567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, M.; Yue, Z.; He, W.; Yang, X.; Yang, G.; Xie, M.; Zhan, D.; Baxter, S.W.; Vasseur, L.; Gurr, G.M.; et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013, 45, 220–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- Lin, L.; Lin, H.; Zhang, M.; Ni, B.; Yin, X.; Qu, C.; Ni, J. A novel method to analyze hepatotoxic components in Polygonum multiflorum using ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry. J. Hazard. Mater. 2015, 299, 249–259. [Google Scholar] [CrossRef]
- Farrés, M.; Platikanov, S.; Tsakovski, S.; Tauler, R. Comparison of the variable importance in projection (VIP) and of the selectivity ratio (SR) methods for variable selection and interpretation. J. Chemom. 2015, 29, 528–536. [Google Scholar] [CrossRef]
- Li, X.Z.; Yang, X.Y.; Wang, Y.; Zhang, S.N.; Zou, W.; Wang, Y.; Li, X.N.; Wang, L.S.; Zhang, Z.G.; Xie, L.Z. Urine metabolic profiling for the pathogenesis research of erosive oral lichen planus. Arch. Oral Biol. 2017, 73, 206–213. [Google Scholar] [CrossRef]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Kindt, R.; Coe, R. Tree Diversity Analysis: A Manual and Software for Common Statistical Methods for Ecological and Biodiversity Studies; World Agroforestry Centre: Nairobi, Kenya, 2005. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ssekagiri, A.; Sloan, W.T.; Ijaz, U.Z. MicrobiomeSeq: An R Package for Microbial Community Analysis in an Environmental Context. 2018. Available online: http://www.github.com/umerijaz/microbiomeSeq (accessed on 10 June 2019).
- Schliep, K.P. Phangorn: Phylogenetic analysis in R. Bioinformatics 2010, 27, 592–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez Arbizu, P. PairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. R Package Version 0.0.1. 2017. Available online: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed on 16 October 2019).
- Love, M.I.; Anders, S.; Huber, W. Differential analysis of count data—The DESeq2 package. Genome Biol. 2014, 15, 10–1186. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frago, E.; Dicke, M.; Godfray, H.C.J. Insect symbionts as hidden players in insect-plant interactions. Trends Ecol. Evol. 2012, 27, 705–711. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects-diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Nikoh, N.; Hosokawa, T.; Oshima, K.; Hattori, M.; Fukatsu, T. Reductive evolution of bacterial genome in insect gut environment. Genome Biol. Evol. 2011, 3, 702–714. [Google Scholar] [CrossRef] [Green Version]
- Eichler, S.; Schaub, G.A. Development of symbionts in triatomine bugs and the effects of infections with trypanosomatids. Exp. Parasitol. 2002, 100, 17–27. [Google Scholar] [CrossRef]
- Wong, A.C.N.; Dobson, A.J.; Douglas, A.E. Gut microbiota dictates the metabolic response of Drosophila to diet. J. Exp. Biol. 2014, 217, 1894–1901. [Google Scholar] [CrossRef] [Green Version]
- Dunbar, H.E.; Wilson, A.C.C.; Ferguson, N.R.; Moran, N.A. Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol. 2007, 5, e96. [Google Scholar] [CrossRef] [PubMed]
- Alborn, H.T.; Turlings, T.C.J.; Jones, T.H.; Stenhagen, G.; Loughrin, J.H.; Tumlinson, J.H. An elicitor of plant volatiles from beet armyworm oral secretion. Science 1997, 276, 945–949. [Google Scholar] [CrossRef]
- Adams, A.S.; Currie, C.R.; Cardoza, Y.; Klepzig, K.D.; Raffa, K.F. Effects of symbiotic bacteria and tree chemistry on the growth and reproduction of bark beetle fungal symbionts. Can. J. For. Res. 2009, 39, 1133–1147. [Google Scholar] [CrossRef]
- Hehemann, J.H.; Correc, G.; Barbeyron, T.; Helbert, W.; Czjzek, M.; Michel, G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 2010, 464, 908–912. [Google Scholar] [CrossRef] [PubMed]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosokawa, T.; Kikuchi, Y.; Shimada, M.; Fukatsu, T. Obligate symbiont involved in pest status of host insect. Proc. R. Soc. B Biol. Sci. 2007, 274, 1979–1984. [Google Scholar] [CrossRef] [Green Version]
- Henry, L.M.; Peccoud, J.; Simon, J.C.; Hadfield, J.D.; Maiden, M.J.C.; Ferrari, J.; Godfray, H.C.J. Horizontally transmitted symbionts and host colonization of ecological niches. Curr. Biol. 2013, 23, 1713–1717. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.M.V.; Huynh, L.Y.; Bolender, C.M.; Nelson, K.G.; McCutcheon, J.P. Population genomics of a symbiont in the early stages of a pest invasion. Mol. Ecol. 2014, 23, 1516–1530. [Google Scholar] [CrossRef]
- Martinez, A.J.; Onchuru, T.O.; Ingham, C.S.; Sandoval-Calderón, M.; Salem, H.; Deckert, J.; Kaltenpoth, M. Angiosperm to Gymnosperm host-plant switch entails shifts in microbiota of the Welwitschia bug, Probergrothius angolensis (Distant, 1902). Mol. Ecol. 2019, 28, 5172–5187. [Google Scholar] [CrossRef]
- Wang, X.Q.; Zhang, A.H.; Miao, J.H.; Sun, H.; Yan, G.L.; Wu, F.F.; Wang, X.J. Gut microbiota as important modulator of metabolism in health and disease. RSC Adv. 2018, 8, 42380–42389. [Google Scholar] [CrossRef] [Green Version]
- Baars, A.; Oosting, A.; Lohuis, M.; Koehorst, M.; El Aidy, S.; Hugenholtz, F.; Smidt, H.; Mischke, M.; Boekschoten, M.V.; Verkade, H.J.; et al. Sex differences in lipid metabolism are affected by presence of the gut microbiota. Sci. Rep. 2018, 8, 13426. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Powell, J.E.; Steele, M.I.; Dietrich, C.; Moran, N.A. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 4775–4780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelgaffar, H.; Tague, E.D.; Castro Gonzalez, H.F.; Campagna, S.R.; Jurat-Fuentes, J.L. Midgut metabolomic profiling of fall armyworm (Spodoptera frugiperda) with field-evolved resistance to Cry1F corn. Insect Biochem. Mol. Biol. 2019, 106, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Talekar, N.S.; Shelton, A.M. Biology, ecology, and management of the diamondback moth. Annu. Rev. Entomol. 1993, 38, 275–301. [Google Scholar] [CrossRef]
- Li, Z.; Feng, X.; Liu, S.-S.; You, M.; Furlong, M.J. Biology, ecology and management of the diamondback moth in China. Annu. Rev. Entomol. 2016, 61, 277–296. [Google Scholar] [CrossRef] [PubMed]
- Furlong, M.J.; Wright, D.J.; Dosdall, L.M. Diamondback Moth ecology and management: Problems, progress, and prospects. Annu. Rev. Entomol. 2013, 58, 517–541. [Google Scholar] [CrossRef]
- Löhr, B.; Gathu, R. Evidence of adaptation of diamonback moth, Plutella xylostella (L.), to pea, Pisum sativum L. Int. J. Trop. Insect Sci. 2002, 22, 161–173. [Google Scholar] [CrossRef]
- Henniges-Janssen, K.; Reineke, A.; Heckel, D.G.; Groot, A.T. Complex inheritance of larval adaptation in Plutella xylostella to a novel host plant. Heredity 2011, 107, 421–432. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.; Sun, B.; Gurr, G.M.; Vasseur, L.; Xue, M.; You, M. Gut microbiota mediate insecticide resistance in the diamondback moth, Plutella xylostella (L.). Front. Microbiol. 2018, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Gupta, P.D.; Thorsteinson, A.J. Food plant relationship of the diamondback moth (Plutella Maculipennis (Curt.)). Entomol. Exp. Appl. 1960, 3, 241–250. [Google Scholar] [CrossRef]
- Dong, H.L.; Zhang, S.X.; Chen, Z.H.; Tao, H.; Li, X.; Qiu, J.F.; Cui, W.Z.; Sima, Y.H.; Cui, W.Z.; Xu, S.Q. Differences in gut microbiota between silkworms (Bombyx mori) reared on fresh mulberry (Morus alba var. multicaulis) leaves or an artificial diet. RSC Adv. 2018, 8, 26188–26200. [Google Scholar] [CrossRef] [Green Version]
- Pinto-Tomás, A.A.; Sittenfeld, A.; Uribe-Lorío, L.; Chavarría, F.; Mora, M.; Janzen, D.H.; Goodman, R.M.; Simon, H.M. Comparison of midgut bacterial diversity in tropical caterpillars (Lepidoptera: Saturniidae) fed on different diets. Environ. Entomol. 2011, 40, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Gurr, G.M.; Vasseur, L.; Zheng, D.; Zhong, H.; Qin, B.; Lin, J.; Wang, Y.; Song, F.; Li, Y.; et al. Metagenomic sequencing of diamondback moth gut microbiome unveils key holobiont adaptations for herbivory. Front. Microbiol. 2017, 8, 663. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.-L.; Xia, X.-F.; Chen, J.-H.; Geoff, M.G.; You, M.-S. Effects of different diets on the diversity of larval gut bacteria of the diamondback moth, Plutella xylostella (Lepidoptera:Plutellidae). Acta Entomol. Sin. 2019, 62, 1172–1185. [Google Scholar]
- Gauthier, J.P.; Outreman, Y.; Mieuzet, L.; Simon, J.C. Bacterial communities associated with host-adapted populations of pea aphids revealed by deep sequencing of 16S ribosomal DNA. PLoS ONE 2015, 10, e0120664. [Google Scholar] [CrossRef]
- Schmid, R.B.; Lehman, R.M.; Brözel, V.S.; Lundgren, J.G. An indigenous gut bacterium, Enterococcus faecalis (Lactobacillales: Enterococcaceae), increases seed consumption by Harpalus pensylvanicus (Coleoptera: Carabidae). Fla. Entomol. 2014, 97, 575–585. [Google Scholar] [CrossRef]
- Yeruva, T.; Vankadara, S.; Ramasamy, S.; Lingaiah, K. Identification of potential probiotics in the midgut of mulberry silkworm, Bombyx mori through metagenomic approach. Probiotics Antimicrob. Proteins 2019, 1–6. [Google Scholar] [CrossRef]
- Sun, Z.; Lu, Y.; Zhang, H.; Kumar, D.; Liu, B.; Gong, Y.; Zhu, M.; Zhu, L.; Liang, Z.; Kuang, S.; et al. Effects of BmCPV infection on silkworm Bombyx mori intestinal bacteria. PLoS ONE 2016, 11, e0146313. [Google Scholar] [CrossRef]
- Chen, B.; Teh, B.S.; Sun, C.; Hu, S.; Lu, X.; Boland, W.; Shao, Y. Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 2016, 6, 29505. [Google Scholar] [CrossRef]
- Van Frankenhuyzen, K. Insecticidal activity of Bacillus thuringiensis crystal proteins. J. Invertebr. Pathol. 2009, 101, 1–16. [Google Scholar] [CrossRef]
- Warnick, T.A.; Methé, B.A.; Leschine, S.B. Clostridium phytofermentans sp. nov., a cellulolytic mesophile from forest soil. Int. J. Syst. Evol. Microbiol. 2002, 52, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- Jenq, R.R.; Taur, Y.; Devlin, S.M.; Ponce, D.M.; Goldberg, J.D.; Ahr, K.F.; Littmann, E.R.; Ling, L.; Gobourne, A.C.; Miller, L.C.; et al. Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol. Blood Marrow Transplant. 2015, 21, 1373–1383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abou-Zaid, M.M.; Beninger, C.W.; Arnason, J.T.; Nozzolillo, C. The effect of one flavone, two catechins and four flavonols on mortality and growth of the European corn borer (Ostrinia nubilalis Hubner). Biochem. Syst. Ecol. 1993, 21, 415–420. [Google Scholar] [CrossRef]
- Pompella, A.; Visvikis, A.; Paolicchi, A.; De Tata, V.; Casini, A.F. The changing faces of glutathione, a cellular protagonist. Biochem. Pharmacol. 2003, 66, 1499–1503. [Google Scholar] [CrossRef]
- Storelli, G.; Defaye, A.; Erkosar, B.; Hols, P.; Royet, J.; Leulier, F. Lactobacillus plantarum promotes drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011, 14, 403–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.-Y.; Saqib, H.S.A.; Chen, J.-H.; Ruan, Q.-Q.; Vasseur, L.; He, W.-Y.; You, M.-S. Differential Profiles of Gut Microbiota and Metabolites Associated with Host Shift of Plutella xylostella. Int. J. Mol. Sci. 2020, 21, 6283. https://doi.org/10.3390/ijms21176283
Yang F-Y, Saqib HSA, Chen J-H, Ruan Q-Q, Vasseur L, He W-Y, You M-S. Differential Profiles of Gut Microbiota and Metabolites Associated with Host Shift of Plutella xylostella. International Journal of Molecular Sciences. 2020; 21(17):6283. https://doi.org/10.3390/ijms21176283
Chicago/Turabian StyleYang, Fei-Ying, Hafiz Sohaib Ahmed Saqib, Jun-Hui Chen, Qian-Qian Ruan, Liette Vasseur, Wei-Yi He, and Min-Sheng You. 2020. "Differential Profiles of Gut Microbiota and Metabolites Associated with Host Shift of Plutella xylostella" International Journal of Molecular Sciences 21, no. 17: 6283. https://doi.org/10.3390/ijms21176283
APA StyleYang, F. -Y., Saqib, H. S. A., Chen, J. -H., Ruan, Q. -Q., Vasseur, L., He, W. -Y., & You, M. -S. (2020). Differential Profiles of Gut Microbiota and Metabolites Associated with Host Shift of Plutella xylostella. International Journal of Molecular Sciences, 21(17), 6283. https://doi.org/10.3390/ijms21176283