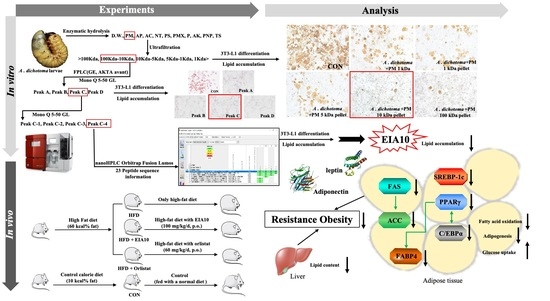
Exploring the Role of a Novel Peptide from Allomyrina dichotoma Larvae in Ameliorating Lipid Metabolism in Obesity
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation, Purification, and Identification of Novel Peptide, EIA10
2.2. Effects of EIA10 on Body Composition in High-Fat Diet (HFD)-Fed Mice
2.3. Effects of EIA10 on Lipid Accumulation and Hepatocellular Damage
2.4. Effects of EIA10 on Blood Biochemical Parameters
2.5. Effects of EIA10 against Diet-Induced Impaired Glucose Homeostasis and Insulin Resistance
2.6. Effects of EIA10 on Lipogenic Molecule Signaling Pathway
3. Materials and Methods
3.1. Biological Materials
3.2. Enzymatic Hydrolysis
3.3. Cell Culture, Differentiation, and Treatments
3.4. Determination of the Deposition of Lipids by Oil Red O Staining
3.5. Fast Protein liquid Chromatography (FPLC)
3.6. Analysis of Target Peptides
3.7. Peptide Synthesis
3.8. Experimental Animals, Diet, and Treatments
3.9. Body Fat Composition Analysis
3.10. Histological Analysis
3.11. Biochemical Analysis
3.12. Oral Glucose Tolerance Test (OGTT) and Insulin Levels
3.13. Estimation of Insulin Resistance Indices
3.14. mRNA Analysis
3.15. Protein Quantification
3.16. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| EIA10 | Glu-Ile-Ala-Gln-Asp-Phe-Lys-Thr-Asp-Leu |
| FAO | Food and Agriculture Organization of the United Nations |
| TG | Triglyceride |
| EPHs | Enzymatic protein hydrolysates |
| Q-TOFMS | Quadrupole time-of-flight mass spectrometry |
| NEFAs | Non-esterified fatty acids |
| AUC | Area under the glucose–tolerance curve |
| FAS | Fatty acid synthase |
| PPARγ | Peroxisomal proliferator-activated receptor γ |
| PNP | Protease NP |
| P | Pancreatin |
| AP | Alphalase NP |
| AK | Alkaline protease |
| PM | Promod 278P |
| AC | Alcalase |
| NT | Neutrase |
| PMX | Protamex |
| FPLC | Fast protein liquid chromatography |
| DMEM | Dulbecco’s modified Eagle medium |
| P/S | Penicillin/streptomycin |
| IEC | Ion-exchange chromatography |
| ACN | Acetonitrile |
| Fmoc | Fluorenylmethylocycarbonyl |
| tBu | tertiary-butyl |
| HPLC | High-performance liquid chromatography |
| LC | Liquid chromatography |
| MS | Mass spectrometry |
| HFD | High-fat diet |
| DXA | Dual-energy X-ray absorptiometry |
| H&E | Hematoxylin and eosin |
| ALT | Alanine aminotransferase |
| AST | Aspartate aminotransferase |
| LDL | Low-density lipoprotein cholesterol |
| HDL | High-density lipoprotein cholesterol |
| ELISA | Enzyme-linked immunosorbent assay |
| OGTT | Oral glucose tolerance test |
| Hepatic IRI | Hepatic insulin resistance index |
| HOMA-IR | Homeostasis model assessment of insulin resistance |
| RT | Reverse transcription |
| PCR | Polymerase chain reaction |
| cDNA | Complementary DNA |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| ANOVA | Analysis of variance |
References
- Nowak, V.; Persijn, D.; Rittenschober, D.; Charrondiere, U.R. Review of food composition data for edible insects. Food Chem. 2016, 193, 39–46. [Google Scholar] [CrossRef]
- Tao, J.; Li, Y.O. Edible insects as a means to address global malnutrition and food insecurity issues. Food Qual. Saf. 2018, 2, 17–26. [Google Scholar] [CrossRef]
- Yamada, M.; Nakamura, K.; Saido-Sakanaka, H.; Asaoka, A.; Yamakawa, M.; Sameshima, T.; Hirota, Y. Effect of modified oligopeptides from the beetle Allomyrina dichotoma on Escherichia coli infection in mice. J. Vet. Med. Sci. 2004, 66, 137–142. [Google Scholar] [CrossRef] [Green Version]
- Ghaly, A.E.; Alkoaik, F.N. The yellow mealworm as a novel source of protein. Am. J. Agric. Biol. Sci. 2009, 4, 319–331. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.S.; Huh, J.; You, G.C.; Chae, S.C.; Lee, O.S.; Lee, H.B.; Lee, G.B.; Kim, J.S. Allomyrina dichotoma larva extracts protect streptozotocin-induced oxidative cytotoxicity. J. Environ. Toxicol. 1986, 22, 349–355. [Google Scholar]
- Chung, M.Y.; Yoon, Y.I.; Hwang, J.S.; Goo, T.W.; Yun, E.Y. Anti-obesity effect of Allomyrina dichotoma (A rthropoda: Insecta) larvae ethanol extract on 3T3-L1 adipocyte differentiation. Entomol. Res. 2014, 44, 9–16. [Google Scholar]
- Kim, H.G.; Kwon, K.; Suh, H.W.; Lee, S.; Park, K.H.; Kwon, O.Y.; Choi, J.Y. Exosome isolation from hemolymph of K orean rhinoceros beetle, Allomyrina dichotoma (Coleoptera: Scarabaeidae). Entomol. Res. 2015, 45, 339–344. [Google Scholar]
- Kim, J.; Yun, E.Y.; Park, S.W.; Goo, T.W.; Seo, M. Allomyrina dichotoma larvae regulate food intake and body weight in high fat diet-induced obese mice through mTOR and Mapk signaling pathways. Nutrients 2016, 8, 100. [Google Scholar] [CrossRef] [Green Version]
- Hu, F.; Zhang, Y.; Song, Y.; Baez, R.V. Lipid metabolism, metabolic syndrome, and cancer. Lipid Metab. 2013, 185–210. [Google Scholar] [CrossRef] [Green Version]
- Ebbert, J.O.; Jensen, M.D. Fat depots, free fatty acids, and dyslipidemia. Nutrients 2013, 5, 498–508. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Chen, X.W.; Huang, X.; Song, B.L.; Wang, Y. Regulation of glucose and lipid metabolism in health and disease. Sci. China Life Sci. 2019, 62, 1–39. [Google Scholar] [CrossRef]
- Johnson, A.A.; Stolzing, A. The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell 2019, 18, e13048. [Google Scholar] [CrossRef] [Green Version]
- Oussaada, S.M.; van Galen, K.A.; Cooiman, M.I.; Kleinendorst, L.; Hazebroek, E.J.; van Haelst, M.M.; Serlie, M.J. The pathogenesis of obesity. Metabolism 2019, 92, 26–36. [Google Scholar] [CrossRef] [Green Version]
- Meldrum, D.R.; Morris, M.A.; Gambone, J.C. Obesity pandemic: Causes, consequences, and solutions—But do we have the will? Fertil. Steril. 2017, 107, 833–839. [Google Scholar] [CrossRef] [Green Version]
- Sumithran, P.; Proietto, J. Benefit-risk assessment of orlistat in the treatment of obesity. Drug Saf. 2014, 37, 597–608. [Google Scholar] [CrossRef]
- Ochiai, M.; MISAkI, K.; Takeuchi, T.; Narumi, R.; Azuma, Y.; Matsuo, T. Egg white hydrolysate can be a low-allergenic food material to suppress ectopic fat accumulation in rats fed an equicaloric diet. J. Nutr. Sci. Vitaminol. 2017, 63, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Garcés-Rimón, M.; González, C.; Uranga, J.A.; López-Miranda, V.; López-Fandiño, R.; Miguel, M. Pepsin egg white hydrolysate ameliorates obesity-related oxidative stress, inflammation and steatosis in Zucker Fatty Rats. PLoS ONE 2016, 11, e0151193. [Google Scholar] [CrossRef] [Green Version]
- C de Campos Zani, S.; Wu, J.; B Chan, C. Egg and soy-derived peptides and hydrolysates: A review of their physiological actions against diabetes and obesity. Nutrients 2018, 10, 549. [Google Scholar] [CrossRef] [Green Version]
- Mudgil, P.; Kamal, H.; Yuen, G.C.; Maqsood, S. Characterization and identification of novel antidiabetic and anti-obesity peptides from camel milk protein hydrolysates. Food Chem. 2018, 259, 46–54. [Google Scholar] [CrossRef]
- Lee, E.J.; Hur, J.; Ham, S.A.; Jo, Y.; Lee, S.; Choi, M.J.; Seo, H.G. Fish collagen peptide inhibits the adipogenic differentiation of preadipocytes and ameliorates obesity in high fat diet-fed mice. Int. J. Biol. Macromol. 2017, 104, 281–286. [Google Scholar] [CrossRef]
- Raksha, N.G.; Potalitsyn, P.Y.; Yurchenko, A.V.; Halenova, T.I.; Savchuk, O.M.; Ostapchenko, L.I. Prevention of diet-induced obesity in rats by oral application of collagen fragments. Arch. Biol. Sci. 2018, 70, 77–86. [Google Scholar] [CrossRef] [Green Version]
- Lemonnier, D. Effect of age, sex, and site on the cellularity of the adipose tissue in mice and rats rendered obese by a high-fat diet. J. Clin. Investig. 1972, 51, 2907–2915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liou, C.J.; Lee, Y.K.; Ting, N.C.; Chen, Y.L.; Shen, S.C.; Wu, S.J.; Huang, W.C. Protective effects of Licochalcone A ameliorates obesity and non-alcoholic fatty liver disease via promotion of the Sirt-1/AMPK pathway in mice fed a high-fat diet. Cells 2019, 8, 447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akcılar, R.; Koçak, F.E.; Şimşek, H.; Akcılar, A.; Bayat, Z.; Ece, E.; Kökdaşgil, H. The effect of adropin on lipid and glucose metabolism in rats with hyperlipidemia. Iran. J. Basic Med. Sci. 2016, 19, 245. [Google Scholar]
- Wu, Y.R.; Shi, X.Y.; Ma, C.Y.; Zhang, Y.; Xu, R.X.; Li, J.J. Liraglutide improves lipid metabolism by enhancing cholesterol efflux associated with ABCA1 and ERK1/2 pathway. Cardiovasc. Diabetol. 2019, 18, 1–12. [Google Scholar] [CrossRef]
- Lv, X.C.; Guo, W.L.; Li, L.; Yu, X.D.; Liu, B. Polysaccharide peptides from Ganoderma lucidum ameliorate lipid metabolic disorders and gut microbiota dysbiosis in high-fat diet-fed rats. J. Funct. Foods 2019, 57, 48–58. [Google Scholar] [CrossRef]
- Orsoni, A.; Thérond, P.; Tan, R.; Giral, P.; Robillard, P.; Kontush, A.; Meikle, P.J.; Chapman, M.J. Statin action enriches HDL3 in polyunsaturated phospholipids and plasmalogens and reduces LDL-derived phospholipid hydroperoxides in atherogenic mixed dyslipidemia. J. Lipid Res. 2016, 57, 2073–2087. [Google Scholar] [CrossRef] [Green Version]
- Peng, C.H.; Liu, L.K.; Chuang, C.M.; Chyau, C.C.; Huang, C.N.; Wang, C.J. Mulberry water extracts possess an anti-obesity effect and ability to inhibit hepatic lipogenesis and promote lipolysis. J. Agric. Food Chem. 2011, 59, 2663–2671. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Cohen, D.E. Triglyceride metabolism in the liver. Compr. Physiol. 2011, 8, 1–22. [Google Scholar]
- Petersen, M.C.; Shulman, G.I. Mechanisms of Insulin Action and Insulin Resistance. Physiol. Rev. 2018, 98, 2133–2223. [Google Scholar] [CrossRef] [Green Version]
- Sharma, B.R.; Kim, H.J.; Rhyu, D.Y. Caulerpa lentillifera extract ameliorates insulin resistance and regulates glucose metabolism in C57BL/KsJ-db/db mice via PI3K/AKT signaling pathway in myocytes. J. Transl. Med. 2015, 13, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Xu, S.; Mihaylova, M.M.; Zheng, B.; Hou, X.; Jiang, B.; Park, O.; Luo, Z.; Lefai, E.; Shyy, J.Y.J.; et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011, 13, 376–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moslehi, A.; Hamidi-zad, Z. Role of SREBPs in liver diseases: A mini-review. J. Clin. Transl. Hepatol. 2018, 6, 332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, S.H.; Wu, K.M.; Guo, Y.Y.; Pickett, J.A.; Field, L.M.; Zhou, J.J.; Zhang, Y.J. Identification of genes expressed in the sex pheromone gland of the black cutworm Agrotis ipsilon with putative roles in sex pheromone biosynthesis and transport. BMC Genomics 2013, 14, 636. [Google Scholar] [CrossRef] [Green Version]
- Carta, G.; Murru, E.; Banni, S.; Manca, C. Palmitic acid: Physiological role, metabolism and nutritional implications. Front. Physiol. 2017, 8, 902. [Google Scholar] [CrossRef] [Green Version]
- Madsen, M.S.; Siersbæk, R.; Boergesen, M.; Nielsen, R.; Mandrup, S. Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading. Mol. Cell. Biol. 2014, 34, 939–954. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Nakajima, T.; Gonzalez, F.J.; Tanaka, N. PPARs as Metabolic Regulators in the Liver: Lessons from Liver-Specific PPAR-Null Mice. Int. J. Mol. Sci. 2020, 21, 2061. [Google Scholar] [CrossRef] [Green Version]
- Park, P.J.; Heo, S.J.; Park, E.J.; Kim, S.K.; Byun, H.G.; Jeon, B.T.; Jeon, Y.J. Reactive oxygen scavenging effect of enzymatic extracts from Sargassum thunbergii. J. Agr. Food Chem. 2005, 53, 6666–6672. [Google Scholar] [CrossRef]
- Rudich, A.; Tirosh, A.; Potashnik, R.; Hemi, R.; Kanety, H.; Bashan, N. Prolonged oxidative stress impairs insulin-induced GLUT4 translocation in 3T3-L1 adipocytes. Diabetes 1998, 47, 1562–1569. [Google Scholar] [CrossRef]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Coccia, F.; Testa, M.; Guarisco, G.; Di Cristofano, C.; Silecchia, G.; Leonetti, F.; Gastaldelli, A.; Capoccia, D. Insulin resistance, but not insulin response, during oral glucose tolerance test (OGTT) is associated to worse histological outcome in obese NAFLD. Nutr. Metab. Cardiovas. 2020, 30, 106–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isokuortti, E.; Zhou, Y.; Peltonen, M.; Bugianesi, E.; Clement, K.; Bonnefont-Rousselot, D.; Lacorte, J.M.; Gastaldelli, A.; Schuppan, D.; Schattenberg, M.; et al. Use of HOMA-IR to diagnose non-alcoholic fatty liver disease: A population-based and inter-laboratory study. Diabetologia 2017, 60, 1873–1882. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Choi, Y.J.; Tang, Y.; Bae, S.M.; Yang, H.P.; Kim, E.K. Efficacy and Mechanism of Polymerized Anthocyanin from Grape-Skin Extract on High-Fat-Diet-Induced Nonalcoholic Fatty Liver Disease. Nutrients 2019, 11, 2586. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.J.; Lee, J.I.; Fan, M.; Tang, Y.; Yoon, E.J.; Ryu, Y.B.; Kim, E.K. Metabolomic Analysis of Morus Cultivar Root Extracts and Their Ameliorative Effect on Testosterone-Induced Prostate Enlargement in Sprague-Dawley Rats. Int. J. Mol. Sci. 2020, 21, 1435. [Google Scholar] [CrossRef] [Green Version]





| No. | Peptide Sequence | Peptide Length | Molecular Composition | Molecular Weight (Da) | Lipid Accumulation (% of Control) |
|---|---|---|---|---|---|
| 1 | KLPFQR (Lys-Leu-Pro-Phe-Gln-Arg) | 6 | C37 H61 N11 O8 | 787 | 57.85 |
| 2 | STELLIR (Ser-Thr-Glu-Leu-Leu-Ile-Arg) | 7 | C36 H66 N10 O12 | 830 | 59.04 |
| 3 | EIAQDFK (Glu-Ile-Ala-Gln-Asp-Phe-Lys) | 7 | C38 H59 N9 O13 | 849 | 52.20 |
| 4 | HLQLAIR (His-Leu-Gln-Leu-Ala-Ile-Arg) | 7 | C38 H67 N13 O9 | 849 | 63.26 |
| 5 | AVQGLLK (Ala-Val-Gln-Gly-Leu-Leu-Lys) | 7 | C33 H61 N9 O9 | 727 | 48.40 |
| 6 | IAQGGVLP (Ile-Ala-Gln-Gly-Gly-Val-Leu-Pro) | 8 | C34 H59 N9 O10 | 753 | 38.01 |
| 7 | NDEELNKLL (Asn-Asp-Glu-Glu-Leu-Asn-Lys-Leu-Leu) | 9 | C46 H78 N12 O18 | 1086 | 39.44 |
| 8 | AGLQFPVGR (Ala-Gly-Leu-Gln-Phe-Pro-Val-Gly-Arg) | 9 | C43 H69 N13 O11 | 943 | 33.70 |
| 9 | YRPGTVALR (Tyr-Arg-Pro-Gly-Thr-Val-Ala-Leu-Arg) | 9 | C46 H77 N15 O12 | 1031 | 40.63 |
| 10 | KSTGGKAPR (Lys-Ser-Thr-Gly-Gly-Lys-Ala-Pro-Arg) | 9 | C37 H68 N14 O12 | 900 | 48.40 |
| 11 | KQLATKAAR (Lys-Gln-Leu-Ala-Thr-Lys-Ala-Ala-Arg) | 9 | C42 H79 N15 O12 | 985 | 94.26 |
| 12 | RFQSSAVMA (Arg-Phe-Gln-Ser-Ser-Ala-Val-Met-Ala) | 9 | C42 H69 N13 O13 S1 | 995 | 57.47 |
| 13 | TLSDYNIQK (Thr-Leu-Ser-Asp-Tyr-Asn-Ile-Gln-Lys) | 9 | C47 H76 N12 O17 | 1080 | 68.00 |
| 14 | NKLLSGVTIA (Asn-Lys-Leu-Leu-Ser-Gly-Val-Thr-Ile-Ala) | 10 | C45 H82 N12 O14 | 1014 | 77.76 |
| * 15 | EIAQDFKTDL (Glu-Ile-Ala-Gln-Asp-Phe-Lys-Thr-Asp-Leu) | 10 | C52 H82 N12 O19 | 1178 | 30.22 |
| 16 | ALNQAWAFLK (Ala-Leu-Asn-Gln-Ala-Trp-Ala-Phe-Leu-Lys) | 10 | C56 H84 N14 O13 | 1160 | 57.47 |
| 17 | AERVGAGAPVY (Ala-Glu-Arg-Val-Gly-Ala-Gly-Ala-Pro-Val-Try) | 11 | C48 H76 N14 O15 | 1088 | 35.06 |
| 18 | RIVDFHMLESR (Arg-Ile-Val-Asp-Phe-His-Met-Leu-Glu-Ser-Arg) | 11 | C61 H99 N19 O17 S1 | 1401 | 41.94 |
| 19 | SGVTIAQGGVLP (Ser-Gly-Val-Thr-Ile-Ala-Gln-Gly-Gly-Val-Leu-Pro) | 12 | C48 H83 N13 O16 | 1097 | 68.00 |
| 20 | APRKQLATKAAR (Ala-Pro-Arg-Lys-Gln-Leu-Ala-Thr-Lys-Ala-Ala-Arg) | 12 | C56 H103 N21 O15 | 1309 | 94.82 |
| 21 | QNIIPASTGAAK (Gln-Asn-Ile-Ile-Pro-Ala-Ser-Thr-Gly-Ala-Ala-Lys) | 12 | C50 H87 N15 O17 | 1169 | 44.40 |
| 22 | HLQLAIRNDEELNK (His-Leu-Gln-Leu-Ala-Ile-Arg-Asn-Asp-Glu-Glu-Leu-Asn-Lys) | 14 | C72 H121 N23 O24 | 1691 | 83.11 |
| 23 | VTIAQGGVLPNIQA (Val-Thr-Ile-Ala-Gln-Gly-Gly-Val-Leu-Phe-Asn-Ile-Gln-Ala) | 14 | C61 H105 N17 O19 | 1379 | 74.36 |
| CON | HFD | HFD+EIA10 | HFD+Orlistat | |
|---|---|---|---|---|
| Initial body weight (g) | 22.61 ± 2.56 | 22.47 ± 0.36 | 23.07 ± 0.33 | 23.55 ± 0.07 |
| Final body weight (g) | 28.41 ± 0.15 | 43.12 ± 0.09 ## | 37.38 ± 2.55 ** | 36.11 ± 3.92 ** |
| Body weight gain (g) | 5.80 ± 0.33 | 20.65 ± 0.92 ## | 12.31 ± 2.54 ** | 12.56 ± 0.45 ** |
| Total energy intake (kcal) | 935.19 ± 43.12 | 1062.21 ± 16.19 ## | 1026.01 ± 2.60 | 1031.35 ± 26.32 |
| Epididymal adipose tissue weight (g) | 0.67 ± 0.17 | 2.19 ± 0.28 ## | 2.06 ± 0.34 ** | 1.90 ± 0.48 ** |
| Subcutaneous adipose tissue weight (g) | 0.66 ± 0.31 | 4.76 ± 0.39 ## | 3.36 ± 0.58 ** | 3.11 ± 0.53 ** |
| Interscapular adipose tissue weight (g) | 0.19 ± 0.05 | 0.26 ± 0.04 # | 0.22 ± 0.06 | 0.26 ± 0.04 * |
| Total fat weight (g) | 3.99 ± 0.71 | 18.15 ± 0.33 ## | 12.71 ± 0.78 ** | 13.71 ± 1.09 ** |
| Liver weight (g) | 1.23 ± 0.27 | 1.72 ± 0.16 ## | 1.22 ± 0.25 ** | 1.14 ± 0.12 ** |
| TG (mg/dL) | 109.44 ± 2.60 | 248.74 ± 4.62 ## | 140.25 ± 5.23 ** | 115.52 ± 4.66 ** |
| TC (mg/dL) | 95.06 ± 2.69 | 132.65 ± 4.52 ## | 107.46 ± 5.15 ** | 113.26 ± 3.40 ** |
| HDL (mg/dL) | 75.81 ± 2.64 | 52.35 ± 3.19 ## | 74.49 ± 2.57 ** | 72.05 ± 4.38 ** |
| LDL/VLDL (mg/dL) | 19.25 ± 3.71 | 80.30 ± 4.12 ## | 32.97 ± 3.42 ** | 41.21 ± 2.59 ** |
| Glucose (mg/dL) | 121.13 ± 8.41 | 175 ± 9.75 ## | 137 ± 11.47 ** | 160.78 ± 13.62 ** |
| ALT (U/L) | 45.38 ± 8.10 | 98.50 ± 13.54 ## | 58.75 ± 13.16 ** | 44.75 ± 8.38 ** |
| AST (U/L) | 110.00 ± 10.90 | 159.63 ± 9.26 ## | 99.20 ± 20.00 ** | 113.88 ± 16.17 ** |
| Abbreviation | Temperature (°C) | pH | Addition (%) | Inactivation | |
|---|---|---|---|---|---|
| Pancreatin | P | 50 | 7 | 1 | 80 °C, 10 min |
| Food Pro Alkaline protease | AK | 50 | 7 | 1 | 80 °C, 10 min |
| Alphalase NP | AP | 50 | 7 | 1 | 80 °C, 10 min |
| Promod 278P | PM | 50 | 7 | 1 | 80 °C, 10 min |
| Alcalase | AC | 50 | 7 | 1 | 80 °C, 10 min |
| Neutrase | NT | 50 | 7 | 1 | 80 °C, 10 min |
| Protamex | PMX | 50 | 7 | 1 | 80 °C, 10 min |
| Protease NP | PNP | 50 | 7 | 1 | 80 °C, 10 min |
| Trypsin | TS | 38 | 7 | 1 | 80 °C, 10 min |
| Pepsin | PS | 38 | 2 (HCl 35%) | 1 | pH 7.5 (NaOH 5N) |
| Control | - | 50 | 7 | - | 80 °C, 10 min |
| Gene Name | Sequence | |
|---|---|---|
| C/EBP-α | Forward | 5-CGT CTA AGA TGA GGG AGT C-3 |
| Reverse | 5-GGC ACA AGG TTA CTT CCT-3 | |
| PPAR-γ | Forward | 5-GAA AGA CAA CGG ACA AAT CAC-3 |
| Reverse | 5-GAA ACT GGC ACC CTT GAA-3 | |
| FAS | Forward | 5-CTT GGG TGC TGA CTA CAA CC-3 |
| Reverse | 5-GCC CTC CCG TAC ACT CAC TC-3 | |
| SREBP-1c | Forward | 5-CTT CTG GAG ACA TCG CAA AC-3 |
| Reverse | 5-GGT AGA CAA CAG CCG CAT C-3 | |
| GAPDH | Forward | 5-GCA CAG TCA AGG CCG AGA AT-3 |
| Reverse | 5-GCC TTC TCC ATG GTG GTG AA-3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bae, S.M.; Fan, M.; Choi, Y.-J.; Tang, Y.; Jeong, G.; Myung, K.; Kim, B.-g.; Kim, E.-K. Exploring the Role of a Novel Peptide from Allomyrina dichotoma Larvae in Ameliorating Lipid Metabolism in Obesity. Int. J. Mol. Sci. 2020, 21, 8537. https://doi.org/10.3390/ijms21228537
Bae SM, Fan M, Choi Y-J, Tang Y, Jeong G, Myung K, Kim B-g, Kim E-K. Exploring the Role of a Novel Peptide from Allomyrina dichotoma Larvae in Ameliorating Lipid Metabolism in Obesity. International Journal of Molecular Sciences. 2020; 21(22):8537. https://doi.org/10.3390/ijms21228537
Chicago/Turabian StyleBae, Sung Mun, Meiqi Fan, Young-Jin Choi, Yujiao Tang, Gwanghui Jeong, Kyungjae Myung, Byung-gyu Kim, and Eun-Kyung Kim. 2020. "Exploring the Role of a Novel Peptide from Allomyrina dichotoma Larvae in Ameliorating Lipid Metabolism in Obesity" International Journal of Molecular Sciences 21, no. 22: 8537. https://doi.org/10.3390/ijms21228537
APA StyleBae, S. M., Fan, M., Choi, Y. -J., Tang, Y., Jeong, G., Myung, K., Kim, B. -g., & Kim, E. -K. (2020). Exploring the Role of a Novel Peptide from Allomyrina dichotoma Larvae in Ameliorating Lipid Metabolism in Obesity. International Journal of Molecular Sciences, 21(22), 8537. https://doi.org/10.3390/ijms21228537