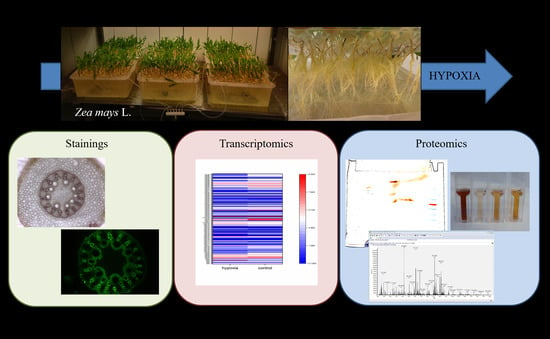
Hypoxia-Responsive Class III Peroxidases in Maize Roots: Soluble and Membrane-Bound Isoenzymes
Abstract
:1. Introduction
2. Results
2.1. RNA Sequence Analyses
2.2. Hydrogen Peroxide Determination, Total Guaiacol Peroxidase Activity and Abundance
2.3. Gel-Free Peroxidase Analyses
2.4. Gel-Based Peroxidase Analyses
2.5. RT-qPCR Analyses
2.6. Root Cross-Sections and In Vivo Root Staining
3. Discussion
3.1. Hypoxia-Responsive Class III Peroxidases
3.2. Membrane Protection and Aerenchyma Formation
3.3. Peroxidase–Rboh Interaction and Cell Wall-Remodeling
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Preparation of Subcellular Fractions
4.3. Hydrogen Peroxide Assay
4.4. Peroxidase Activity
4.5. Gel-Based Analyses and Mass Spectrometry
4.6. Gel-Free Peroxidase Analyses
4.7. Isolation of Total RNA
4.8. Quality Control and RNA Sequencing (RNA Seq)
4.9. Quantitative Reverse-Transcription Polymerase Chain Reaction (RT-qPCR)
4.10. In Vivo Cell Wall Staining
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 2D | two-dimensional |
| ACN | acetonitrile |
| At | Arabidopsis thaliana |
| BSA | bovine serum albumin |
| C | control sample |
| CAD | cinnamyl alcohol dehydrogenase |
| CCoAOMT | caffeoyl-CoA O-methyltransferase 1 |
| cDNA | copy desoxyribonucleic acid |
| CESA | cellulose synthase |
| CET | Central European Time |
| CHAPS | 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate |
| co | Cortex |
| Cox2 | cytochrome c oxidase |
| CSE | caffeoylshikimate esterase |
| Cv | Cultivar |
| DEGs | differentially expressed genes |
| DIR | Dirigent |
| ε470 nm | extinction coefficient at 470 nm |
| ECL | enhanced chemiluminescence |
| EDTA | ethylenediaminetetraacetic acid |
| EF | elongation factor |
| en | Endodermis |
| ER | endoplasmic reticulum |
| ex | Exodermis |
| EXP | Expansin |
| EXPL | expansin-like |
| FCA | fuchsin, chrysoidin, and Astra blue |
| FDR | false discovery rate |
| FLA | fasciclin-like arabinogalactan |
| FPKM | fragments per kilobase million |
| GDPD | glycerophosphodiesterase |
| GDPDL | glycerophosphodiester phosphodiesterase-like |
| H | hypoxia-stressed sample |
| H+-ATPase | PM specific H+ATPase |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| HHT | hydroxycinnamoyl-CoA:ω-hydroxyacid O-hydroxycinnamoyltransferase |
| hy | hypodermis |
| IEF | isoelectric focusing |
| IntDen/area | Integrated Density per area |
| kDa | kilodalton |
| LC–MS/MS | liquid chromatography mass spectrometry |
| LFQ | label-free quantifications |
| micro | microsomes |
| MS | mass spectrometry |
| MW | molecular weight |
| NADH | nicotinamide adenine dinucleotide |
| NCBI | National Center for Biotechnology Information |
| PAGE | polyacrylamid gel electrophoresis |
| ph | phloem |
| pI | point isoelectric |
| PM | plasma membrane |
| ppm | parts per million |
| Prx | class III peroxidases |
| QC | quality control |
| Rboh | respiratory burst oxidase homologs |
| RIN | RNA integrity number |
| RNA | ribonucleic acid |
| RNA Seq | RNA sequence analyses |
| ROL | radial oxygen loss |
| ROS | reactive oxygen species |
| RT | room temperature |
| RT-qPCR | real-time quantitative polymerase chain reaction; quantitative reverse-transcription polymerase chain reaction |
| SB12 | n-dodecyl-N, N-dimethyl-3-ammonio-1-propanesulfonate |
| sc | vascular sclerenchyma cells |
| SDS | sodiumdodecylsulfate |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
| TAE | tris-acetate-EDTA |
| TMB | 3,3′,5,5′-Tetramethylbenzidine |
| Tris | tris(hydroxymethyl)aminomethane |
| Triton X-100 | 2-[4-(2,4,4-trimethylpentan-2-yl)phenoxyl] ethanol |
| tufM | thermo unstable translation elongation factor, mitochondrial |
| V-PPase | pyrophosphate-energized vacuolar membrane proton pump 1 |
| xy | xylem |
| Zm | Zea mays |
References
- Bailey-Serres, J.; Lee, S.C.; Brinton, E. Waterproofing crops: Effective flooding survival strategies. Plant. Physiol. 2012, 160, 1698–1709. [Google Scholar] [CrossRef] [Green Version]
- Nishiuchi, S.; Yamauchi, T.; Takahashi, H.; Kotula, L.; Nakazono, M. Mechanisms for coping with submergence and waterlogging in rice. Rice 2012, 5, 2. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, S.; Shirasaka, N.; Sakata, K.J. ‘Omics’ techniques for identifying flooding-response mechanisms in soybean. J. Proteom. 2013, 93, 169–178. [Google Scholar] [CrossRef]
- Yamauchi, T.; Shimamura, S.; Nakazono, M.; Mochizuki, T. Aerenchyma formation in crop species: A review. Field Crops Res. 2013, 152, 8–16. [Google Scholar] [CrossRef]
- Yordanova, R.Y.; Popova, L.P. Flooding-induced changes in photosynthesis and oxidative status in maize plants. Acta Physiol. Plant. 2007, 29, 535–541. [Google Scholar] [CrossRef]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. 2003, 91, 179–194. [Google Scholar] [CrossRef] [Green Version]
- Noctor, G.; Reichheld, J.-P.; Foyer, C.H. ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. 2018, 80, 3–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smirnoff, N.; Arnaud, D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef] [PubMed]
- Monk, L.S.; Fagerstedt, K.V.; Crawford, R.M.M. Oxygen toxicity and superoxide dismutase as an antioxidant in physiological stress. Physiol. Plant 1989, 76, 456–459. [Google Scholar] [CrossRef]
- Yu, Q.; Rengel, Z. Drought and salinity differentially influence activities of superoxide dismutase in narrow-leafed lupins. Plant Sci. 1999, 142, 1–11. [Google Scholar] [CrossRef]
- Gunawardena, A.H.L.A.N.; Pearce, D.M.E.; Jackson, M.B.; Hawes, C.R.; Evans, D.E. Rapid changes in cell wall pectic polysaccharides are closely associated with early stages of aerenchyma formation, a spatially localized form of programmed cell death in roots of maize (Zea mays L.) promoted by ethylene. Plant Cell Environ. 2001, 24, 1369–1375. [Google Scholar] [CrossRef] [Green Version]
- Arora, K.; Panda, K.K.; Mittal, S.; Mallikarjuna, M.G.; Rao, A.R.; Dash, P.K.; Thirunavukkarasu, N. RNAseq revealed the important gene pathways controlling adaptive mechanisms under waterlogged stress in maize. Sci. Rep. 2017, 7, 10950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajhi, I.; Yamauchi, T.; Takahashi, H.; Nishiuchi, S.; Shiono, K.; Watanabe, R.; Mliki, A.; Nagamura, Y.; Tsutsumi, N.; Nishizawa, N.K.; et al. Identification of genes expressed in maize root cortical cells during lysigenous aerenchyma formation using laser microdissection and microarray analyses. New Phytol. 2011, 190, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Safavi-Rizi, V.; Herde, M.; Stöhr, C. RNA-Seq reveals novel genes and pathways associated with hypoxia duration and tolerance in tomato root. Sci. Rep. 2020, 10, 1692. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Nishiuchi, S.; Kulichikhin, K.; Nakazono, M. Does suberin accumulation in plant roots contribute to waterlogging tolerance? Front. Plant. Sci. 2013, 4, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abiko, T.; Kotula, L.; Shiono, K.; Malik, A.I.; Colmer, T.D.; Nakazono, M. Enhanced formation of aerenchyma and induction of a barrier to radial oxygen loss in adventitious roots of Zea nicaraguensis contribute to its waterlogging tolerance as compared with maize (Zea mays ssp. mays). Plant. Cell Environ. 2012, 35, 1618–1630. [Google Scholar] [CrossRef] [PubMed]
- Castillo, F. Extracellular peroxidases as markers of stress. In Molecular and Physiological Aspects of Plant. Peroxidases; Greppin, H., Penel, C., Gaspar, T., Eds.; Université de Genève, Centre de botanique: Geneva, Switzerland, 1986; pp. 419–426. [Google Scholar]
- Komatsu, S.; Hiraga, S.; Yanagawa, Y. Proteomics techniques for the development of flood tolerant crops. J. Proteome Res. 2012, 11, 68–78. [Google Scholar] [CrossRef]
- Mika, A.; Boenisch, M.J.; Hopff, D.; Lüthje, S. Membrane-bound guaiacol peroxidases from maize (Zea mays L.) roots are regulated by methyl jasmonate, salicylic acid, and pathogen elicitors. J. Exp. Bot. 2010, 61, 831–841. [Google Scholar] [CrossRef] [Green Version]
- Meisrimler, C.N.; Buck, F.; Lüthje, S. Alterations in soluble class III peroxidases of maize shoots by flooding stress. Proteomes 2014, 2, 303–322. [Google Scholar] [CrossRef] [Green Version]
- Thirunavukkarasu, N.; Hossain, F.; Mohan, S.; Shiriga, K.; Mittal, S.; Sharma, R.; Singh, R.K.; Gupta, H.S. Genome-wide expression of transcriptomes and their co-expression pattern in subtropical maize (Zea mays L.) under waterlogging stress. PLoS ONE 2013, 8, e70433. [Google Scholar] [CrossRef] [Green Version]
- De Gara, L. Class III peroxidases and ascorbate metabolism in plants. Phytochem. Rev. 2004, 3, 195–205. [Google Scholar] [CrossRef]
- Lüthje, S.; Meisrimler, C.N.; Hopff, D.; Möller, B. Phylogeny, topology, structure and functions of membrane-bound class III peroxidases in vascular plants. Phytochemistry 2011, 72, 1124–1135. [Google Scholar] [CrossRef] [PubMed]
- Cosio, C.; Dunand, C. Specific functions of individual class III peroxidase genes. J. Exp. Bot. 2009, 60, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Cosio, C.; Penel, C.; Dunand, C. Peroxidases have more functions than a Swiss army knife. Plant. Cell Rep. 2005, 24, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [Green Version]
- Vishwanath, S.J.; Delude, C.; Domergue, F.; Rowland, O. Suberin: Biosynthesis, regulation, and polymer assembly of a protective extracellular barrier. Plant. Cell Rep. 2015, 34, 573–586. [Google Scholar] [CrossRef]
- Novo-Uzal, E.; Fernandez-Perez, F.; Herrero, J.; Gutierrez, J.; Gomez-Ros, L.V.; Bernal, M.A.; Diaz, J.; Cuello, J.; Pomar, F.; Pedreno, M.A. From Zinnia to Arabidopsis: Approaching the involvement of peroxidases in lignification. J. Exper. Bot. 2013, 64, 3499–3518. [Google Scholar] [CrossRef] [Green Version]
- Almagro, L.; Gomez Ros, L.V.; Belchi-Navarro, S.; Bru, R.; Ros Barcelo, A.; Pedreno, M.A. Class III peroxidases in plant defence reactions. J. Exp. Bot. 2009, 60, 377–390. [Google Scholar] [CrossRef] [Green Version]
- Fry, S.C. Oxidative coupling of tyrosine and ferulic acid residues: Intra- andextra-protoplasmic occurrence, predominance of trimers and larger products, and possible role in inter-polymeric cross-linking. Phytochem. Rev. 2004, 3, 97–111. [Google Scholar] [CrossRef]
- Martinez-Rubio, R.; Acebes, J.L.; Encina, A.; Karkonen, A. Class III peroxidases in cellulose deficient cultured maize cells during cell wall remodeling. Physiol. Plant. 2018, 164, 45–55. [Google Scholar] [CrossRef]
- Sekhon, R.S.; Lin, H.; Childs, K.L.; Hansey, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Genome-wide atlas of transcription during maize development. Plant. J. 2011, 66, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Q.; Zhao, Y.; Han, G.; Zhu, S. Systematic analysis of maize class III peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 2015, 566, 95–108. [Google Scholar] [CrossRef]
- Mika, A.; Buck, F.; Lüthje, S. Membrane-bound class III peroxidases: Identification, biochemical properties and sequence analysis of isoenzymes purified from maize (Zea mays L.) roots. J. Proteom. 2008, 71, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Mika, A.; Lüthje, S. Properties of guaiacol peroxidase activities isolated from corn root plasma membranes. Plant. Physiol. 2003, 132, 1489–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lüthje, S.; Martinez-Cortes, T. Membrane-bound class III peroxidases: Unexpected enzymes with exciting functions. Int. J. Mol. Sci. 2018, 19, 2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pike, L.J. Lipid rafts heterogeneity on the high seas. Biochem. J. 2004, 378, 281–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Everaert, J.; Podina, I.R.; Koster, E.H.W. A comprehensive meta-analysis of interpretation biases in depression. Clin. Psychol. Rev. 2017, 58, 33–48. [Google Scholar] [CrossRef] [Green Version]
- Liszkay, A.; Kenk, B.; Schopfer, P. Evidence for the involvement of cell wall peroxidase in the generation of hydroxyl radicals mediating extension growth. Planta 2003, 217, 658–667. [Google Scholar] [CrossRef]
- Lee, Y.; Rubio, M.C.; Alassimone, J.; Geldner, N. A mechanism for localized lignin deposition in the endodermis. Cell 2013, 153, 402–412. [Google Scholar] [CrossRef] [Green Version]
- Ito, S.; Suzuki, Y.; Miyamoto, K.; Ueda, J.; Yamaguchi, I. AtFLA11, a fasciclin-like arabinogalactan-protein, specifically localized in sclerenchyma cells. Biosci. Biotechnol. Biochem. 2005, 69, 1963–1969. [Google Scholar] [CrossRef]
- Von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, N.H.; Selvaraj, G.; Wei, Y.; King, J. Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 2009, 60, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; Ishii, T.; Matsunaga, T.; Tominaga, R.; Kuromori, T.; Wada, T.; Shinozaki, K.; Hirayama, T. The glycerophosphoryl diester phosphodiesterase-like proteins SHV3 and its homologs play important roles in cell wall organization. Plant Cell Physiol. 2008, 49, 1522–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santiago, R.; Barros-Rios, J.; Malvar, R.A. Impact of cell wall composition on maize resistance to pests and diseases. Int. J. Mol. Sci. 2013, 14, 6960–6980. [Google Scholar] [CrossRef] [Green Version]
- Encina, A.; Fry, S.C. Oxidative coupling of a feruloyl-arabinoxylan trisaccharide (FAXX) in the walls of living maize cells requires endogenous hydrogen peroxide and is controlled by a low-Mr apoplastic inhibitor. Planta 2005, 223, 77–89. [Google Scholar] [CrossRef]
- Roberts, E.; Kutchan, T.; Kolattukudy, P.E. Cloning and sequencing of cDNA for a highly anionic peroxidase from potato and the induction of its mRNA in suberizing potato tubers and tomato fruits. Plant. Mol. Biol. 1988, 11, 15–26. [Google Scholar] [CrossRef]
- Whetten, R.W.; MacKay, J.J.; Sederoff, R.R. Recent advantages in understanding lignin biosynthesis. Ann. Rev. Plant. Physiol. Plant. Mol. Biol. 1998, 49, 585–609. [Google Scholar] [CrossRef]
- Enstone, D.; Peterson, C.A. Suberin lamella development in maize seedling roots grown in aerated and stagnant conditions. Plant Cell. Environ. 2006, 28, 444–455. [Google Scholar] [CrossRef]
- Tylova, E.; Peckova, E.; Blascheova, Z.; Soukup, A. Casparian bands and suberin lamellae in exodermis of lateral roots: An important trait of roots system response to abiotic stress factors. Ann. Bot. 2017, 120, 71–85. [Google Scholar] [CrossRef] [Green Version]
- Luthje, S.; Meisrimler, C.N.; Hopff, D.; Schutze, T.; Koppe, J.; Heino, K. Class III peroxidases. Methods Mol. Biol. 2014, 1072, 687–706. [Google Scholar] [CrossRef]
- Bradford, M.M. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Gay, C.; Collins, J.; Gebicki, J.M. Hydroperoxide assay with the ferric-xylenol orange complex. Anal. Biochem. 1999, 273, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Gay, C.A.; Gebicki, J.M. Perchloric acid enhances sensitivity and reproducibility of the ferric-xylenol orange peroxide assay. Anal. Biochem. 2002, 304, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Sturmer, L.R.; Dodd, D.; Chao, C.S.; Shi, R.Z. Clinical utility of an ultrasensitive late night salivary cortisol assay by tandem mass spectrometry. Steroids 2018, 129, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Hopff, D.; Wienkoop, S.; Luthje, S. The plasma membrane proteome of maize roots grown under low and high iron conditions. J. Proteom. 2013, 91, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Turetschek, R.; Desalegn, G.; Epple, T.; Kaul, H.P.; Wienkoop, S. Key metabolic traits of Pisum sativum maintain cell vitality during Didymella pinodes infection: Cultivar resistance and the microsymbionts’ influence. J. Proteom. 2017, 169, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Weckwerth, W. Covain: A toolbox for uni- and multivariate statistics, time-series and correlation network analysis and inverse estimation of the differential Jacobian from metabolomics covariance data. Metabolomics 2012, 8, 81–93. [Google Scholar] [CrossRef]
- Yamashita, D.; Kimura, S.; Wada, M.; Takabe, K. Improved Mäule color reaction provides more detailed information on syringyl lignin distribution in hardwood. J. Wood Sci. 2016, 62, 131–137. [Google Scholar] [CrossRef] [Green Version]
- Chapple, C.C.S.; Vogt, T.; Ellis, B.E.; Somerville, C.R. An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant. Cell 1992, 4, 1413–1424. [Google Scholar]
- Franke, R.; McMichael, C.M.; Meyer, K.; Shirley, A.M.; Cusumano, J.C.; Chapple, C. Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 2000, 22, 223–234. [Google Scholar] [CrossRef] [Green Version]
- Leitner, A.; Lechner, H.; Peter, K. Colour Tests for Precursor Chemicals of Amphetamine-Type Substances; United Nations, Office of Drugs and Crime: Vienna, Austria, 2007. [Google Scholar]
- Crocker, E.C. An experimental study of the significance of “lignin” color reactions. J. Industr. Engineer. Chem. 1921, 13, 625–627. [Google Scholar] [CrossRef]
- Turrell, F.M.; Fisher, P.L. The proximate chemical constituents of citrus woods, with special reference to lignin. Plant Physiol. 1942, 17, 558–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brundrett, M.C.; Enstone, D.E.; Peterson, C.A. A berberine-aniline blue fluorescent stainingprocedure for suberin, lignin and callose in plant tissue. Protoplasma 1988, 146, 133–142. [Google Scholar] [CrossRef]
- Ranathunge, K.; Kim, Y.X.; Wassmann, F.; Kreszies, T.; Zeisler, V.; Schreiber, L. The composite water and solute transport of barley (Hordeum vulgare) roots: Effect of suberized barriers. Ann. Bot. 2017, 119, 629–643. [Google Scholar] [CrossRef] [Green Version]
- Peng, T.; Thorn, K.; Schroeder, T.; Wang, L.; Theis, F.J.; Marr, C.; Navab, N. A BaSiC tool for background and shading correction of optical microscopy images. Nat. Commun. 2017, 8, 14836. [Google Scholar] [CrossRef]








| Name of Peroxidase | Forward Primer | Reverse Primer |
|---|---|---|
| zmprx01 | ACTTGTTCAAGGCCAAGGAG | TTCGTGCTTGTGTTCCAGAC |
| zmprx03 | TCAAGATGGGGCAGATCGAG | ACTCCAGTGAATCCTGATGGG |
| zmprx24 | GGCTCATCCGCATCTTCTT | TGGTTGGGTACCTCGATCT |
| zmprx66 | CGACATGGTTGCACTCTCAG | CGAAGGCGGAGTTGATGTTG |
| zmprx70 | CCACCTCCATGACTGCTTTG | TTCGGATTAGCGGTCTGCTC |
| zmprx81 | CAGGAGGATGACTTCGCCAG | CCGTTGTAGGGTCCCTGATG |
| zmprx85 | GACGCTGAGGAAGAACAAGG | CTGGTCGAAGAACCACCAG |
| zmtufM | CGCAGTTGATGAGTACATCC | AACACGCCCAGTAACAACAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hofmann, A.; Wienkoop, S.; Harder, S.; Bartlog, F.; Lüthje, S. Hypoxia-Responsive Class III Peroxidases in Maize Roots: Soluble and Membrane-Bound Isoenzymes. Int. J. Mol. Sci. 2020, 21, 8872. https://doi.org/10.3390/ijms21228872
Hofmann A, Wienkoop S, Harder S, Bartlog F, Lüthje S. Hypoxia-Responsive Class III Peroxidases in Maize Roots: Soluble and Membrane-Bound Isoenzymes. International Journal of Molecular Sciences. 2020; 21(22):8872. https://doi.org/10.3390/ijms21228872
Chicago/Turabian StyleHofmann, Anne, Stefanie Wienkoop, Sönke Harder, Fabian Bartlog, and Sabine Lüthje. 2020. "Hypoxia-Responsive Class III Peroxidases in Maize Roots: Soluble and Membrane-Bound Isoenzymes" International Journal of Molecular Sciences 21, no. 22: 8872. https://doi.org/10.3390/ijms21228872
APA StyleHofmann, A., Wienkoop, S., Harder, S., Bartlog, F., & Lüthje, S. (2020). Hypoxia-Responsive Class III Peroxidases in Maize Roots: Soluble and Membrane-Bound Isoenzymes. International Journal of Molecular Sciences, 21(22), 8872. https://doi.org/10.3390/ijms21228872