A Pilot Study on the Effects of l-Carnitine and Trimethylamine-N-Oxide on Platelet Mitochondrial DNA Methylation and CVD Biomarkers in Aged Women
Abstract
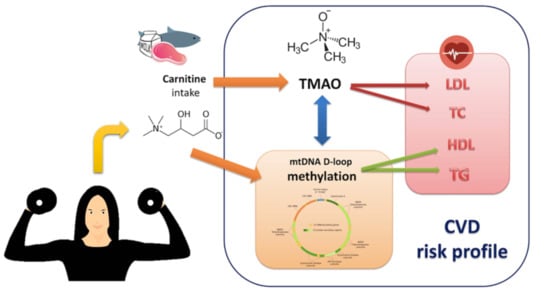
:1. Background
2. Results
2.1. Descriptive Statistics of Body Composition and TMAO Levels before Intervention
2.2. Body Composition Parameters and Age Did Not Affect Basal TMAO Levels
2.3. l-carnitine Supplementation Increased TMAO Levels after 6 Months of Supplementation and Physical Training
2.4. Lipid Profile after 6 Months of Supplementation and Training
2.5. TMAO Levels after 6 Months Correlated with TC and LDL-c
2.6. MtDNA Methylation Levels Analysis at MTCO1 and the D-Loop Region
2.7. D-Loop Methylation Inversely Correlated with TC and LDL in the Total Group of Subjects after 6 Months of Intervention
2.8. Increased D-Loop Methylation Was Associated with a Better Lipid Profile for HDL and TG in the l-leucine + l-carnitine Group
2.9. Prediction Models for TMAO and D-Loop Methylation Increase
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Human Experimental Protocol
4.3. Resistance Exercise Training
4.4. Supplementation
4.5. Samples Collection
4.6. Body Composition Assessment
4.7. TMAO Assessment
4.8. Lipid Profile Assessment
4.9. DNA Extraction and Bisulphite Conversion
4.10. Measurement of mtDNA Methylation
4.11. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 1RM | One-repetition maximum |
| CVD | Cardiovascular disease |
| HDL-c | High-density lipoprotein cholesterol |
| LDL-c | Low-density lipoprotein cholesterol |
| MTCO1 | Mitochondrial cytochrome c oxidase subunit 1 gene |
| mtDNA | Mitochondrial DNA |
| ROC | Receiver operating characteristic |
| TC | Total cholesterol |
| TG | Triglycerides |
| TMAO | Trimethylamine-N-Oxide |
References
- Lamelas, P.M.; Yusuf, S.; Schwalm, J.D. Effective approaches to address the global cardiovascular disease burden. Curr. Opin. Cardiol. 2017, 32, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Rippe, J.M. Lifestyle Strategies for Risk Factor Reduction, Prevention, and Treatment of Cardiovascular Disease. Am. J. Lifestyle Med. 2019, 13, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidou, V.; Daimiel, L.; Ordovas, J.M. Personalized nutrition and cardiovascular disease prevention: From Framingham to PREDIMED. Adv. Nutr. 2014, 5, 368S–371S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loche, E.; Ozanne, S.E. Early nutrition, epigenetics, and cardiovascular disease. Curr. Opin. Lipidol. 2016, 27, 449–458. [Google Scholar] [CrossRef] [Green Version]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.H.W.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.-M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Rossner, R.; Kaeberlein, M.; Leiser, S.F. Flavin-containing monooxygenases in aging and disease: Emerging roles for ancient enzymes. J. Biol. Chem. 2017, 292, 11138–11146. [Google Scholar] [CrossRef] [Green Version]
- Heianza, Y.; Ma, W.; Manson, J.E.; Rexrode, K.M.; Qi, L. Gut Microbiota Metabolites and Risk of Major Adverse Cardiovascular Disease Events and Death: A Systematic Review and Meta-Analysis of Prospective Studies. J. Am. Heart Assoc. 2017, 6, e004947. [Google Scholar] [CrossRef]
- Schiattarella, G.G.; Sannino, A.; Toscano, E.; Giugliano, G.; Gargiulo, G.; Franzone, A.; Trimarco, B.; Esposito, G.; Perrino, C. Gut microbe-generated metabolite trimethylamine-N-oxide as cardiovascular risk biomarker: A systematic review and dose-response meta-analysis. Eur. Heart J. 2017, 38, 2948–2956. [Google Scholar] [CrossRef] [Green Version]
- Canyelles, M.; Tondo, M.; Cedó, L.; Farràs, M.; Escolà-Gil, J.C.; Blanco-Vaca, F. Trimethylamine N-Oxide: A Link among Diet, Gut Microbiota, Gene Regulation of Liver and Intestine Cholesterol Homeostasis and HDL Function. Int. J. Mol. Sci. 2018, 19, 3228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Jiao, X.; Ma, Y.; Liu, Y.; Zhang, L.; He, Y.; Chen, Y. Trimethylamine N-oxide induces inflammation and endothelial dysfunction in human umbilical vein endothelial cells via activating ROS-TXNIP-NLRP3 inflammasome. Biochem. Biophys. Res. Commun. 2016, 481, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Obeid, R.; Awwad, H.M.; Rabagny, Y.; Graeber, S.; Herrmann, W.; Geisel, J. Plasma trimethylamine N-oxide concentration is associated with choline, phospholipids, and methyl metabolism. Am. J. Clin. Nutr. 2016, 103, 703–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.; Gregory, J.C.; Org, E.; Buffa, J.A.; Gupta, N.; Wang, Z.; Li, L.; Fu, X.; Wu, Y.; Mehrabian, M.; et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell 2016, 165, 111–124. [Google Scholar] [CrossRef] [Green Version]
- Baccarelli, A.A.; Byun, H.-M. Platelet mitochondrial DNA methylation: A potential new marker of cardiovascular disease. Clin. Epigenetics 2015, 7, 44. [Google Scholar] [CrossRef] [Green Version]
- Muka, T.; Koromani, F.; Portilla, E.; O’Connor, A.; Bramer, W.M.; Troup, J.; Chowdhury, R.; Dehghan, A.; Franco, O.H. The role of epigenetic modifications in cardiovascular disease: A systematic review. Int. J. Cardiol. 2016, 212, 174–183. [Google Scholar] [CrossRef] [Green Version]
- Aavik, E.; Babu, M.; Yla-Herttuala, S. DNA methylation processes in atheosclerotic plaque. Atherosclerosis 2019, 281, 168–179. [Google Scholar] [CrossRef]
- Voelter-Mahlknecht, S. Epigenetic associations in relation to cardiovascular prevention and therapeutics. Clin. Epigenetics 2016, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Chistiakov, D.A.; Orekhov, A.N.; Bobryshev, Y.V. Treatment of cardiovascular pathology with epigenetically active agents: Focus on natural and synthetic inhibitors of DNA methylation and histone deacetylation. Int. J. Cardiol. 2017, 227, 66–82. [Google Scholar] [CrossRef]
- Metzinger, L.; de Franciscis, S.; Serra, R. The Management of Cardiovascular Risk through Epigenetic Biomarkers. Biomed Res. Int. 2017, 2017, 9158572. [Google Scholar] [CrossRef] [Green Version]
- Costantino, S.; Libby, P.; Kishore, R.; Tardif, J.-C.; El-Osta, A.; Paneni, F. Epigenetics and precision medicine in cardiovascular patients: From basic concepts to the clinical arena. Eur. Heart J. 2018, 39, 4150–4158. [Google Scholar] [CrossRef]
- Lambertini, L.; Byun, H.-M. Mitochondrial Epigenetics and Environmental Exposure. Curr. Environ. Heal. Rep. 2016, 3, 214–224. [Google Scholar] [CrossRef]
- Minocherhomji, S.; Tollefsbol, T.O.; Singh, K.K. Mitochondrial regulation of epigenetics and its role in human diseases. Epigenetics 2012, 7, 326–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castegna, A.; Iacobazzi, V.; Infantino, V. The mitochondrial side of epigenetics. Physiol. Genom. 2015, 47, 299–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bordoni, L.; Smerilli, V.; Nasuti, C.; Gabbianelli, R. Mitochondrial DNA methylation and copy number predict body composition in a young female population. J. Transl. Med. 2019, 17, 399. [Google Scholar] [CrossRef] [PubMed]
- Stimpfel, M.; Jancar, N.; Virant-Klun, I. New Challenge: Mitochondrial Epigenetics? Stem Cell Rev. 2018, 14, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Mposhi, A.; Van der Wijst, M.G.; Faber, K.N.; Rots, M.G. Regulation of mitochondrial gene expression, the epigenetic enigma. Front. Biosci. Landmark Ed. 2017, 22, 1099–1113. [Google Scholar] [PubMed] [Green Version]
- Ferreira, A.; Serafim, T.L.; Sardao, V.A.; Cunha-Oliveira, T. Role of mtDNA-related mitoepigenetic phenomena in cancer. Eur. J. Clin. Invest. 2015, 45 (Suppl. 1), 44–49. [Google Scholar] [CrossRef]
- Coppedè, F.; Stoccoro, A. Mitoepigenetics and Neurodegenerative Diseases. Front. Endocrinol. (Lausanne) 2019, 10, 86. [Google Scholar] [CrossRef] [Green Version]
- D’Aquila, P.; Bellizzi, D.; Passarino, G. Mitochondria in health, aging and diseases: The epigenetic perspective. Biogerontology 2015, 16, 569–585. [Google Scholar] [CrossRef]
- Zheng, L.D.; Linarelli, L.E.; Brooke, J.; Smith, C.; Wall, S.S.; Greenawald, M.H.; Seidel, R.W.; Estabrooks, P.A.; Almeida, F.A.; Cheng, Z. Mitochondrial Epigenetic Changes Link to Increased Diabetes Risk and Early-Stage Prediabetes Indicator. Oxid. Med. Cell. Longev. 2016, 2016, 5290638. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.D.; Linarelli, L.E.; Liu, L.; Wall, S.S.; Greenawald, M.H.; Seidel, R.W.; Estabrooks, P.A.; Almeida, F.A.; Cheng, Z. Insulin resistance is associated with epigenetic and genetic regulation of mitochondrial DNA in obese humans. Clin. Epigenetics 2015, 7, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janeiro, M.H.; Ramirez, M.J.; Milagro, F.I.; Martinez, J.A.; Solas, M. Implication of Trimethylamine N-Oxide (TMAO) in Disease: Potential Biomarker or New Therapeutic Target. Nutrients 2018, 10, e1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowinski, A.; Ufnal, M. Trimethylamine N-oxide: A harmful, protective or diagnostic marker in lifestyle diseases? Nutrition 2018, 46, 7–12. [Google Scholar] [CrossRef]
- Samulak, J.J.; Sawicka, A.K.; Hartmane, D.; Grinberga, S.; Pugovics, O.; Lysiak-Szydlowska, W.; Olek, R.A. L-Carnitine Supplementation Increases Trimethylamine-N-Oxide but not Markers of Atherosclerosis in Healthy Aged Women. Ann. Nutr. Metab. 2019, 74, 11–17. [Google Scholar] [CrossRef]
- Wang, Z.-Y.; Liu, Y.-Y.; Liu, G.-H.; Lu, H.-B.; Mao, C.-Y. l-Carnitine and heart disease. Life Sci. 2018, 194, 88–97. [Google Scholar] [CrossRef]
- Ferrari, R.; Merli, E.; Cicchitelli, G.; Mele, D.; Fucili, A.; Ceconi, C. Therapeutic effects of L-carnitine and propionyl-L-carnitine on cardiovascular diseases: A review. Ann. N. Y. Acad. Sci. 2004, 1033, 79–91. [Google Scholar] [CrossRef]
- Song, X.; Qu, H.; Yang, Z.; Rong, J.; Cai, W.; Zhou, H. Efficacy and Safety of L-Carnitine Treatment for Chronic Heart Failure: A Meta-Analysis of Randomized Controlled Trials. Biomed Res. Int. 2017, 2017, 6274854. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Levison, B.S.; Hazen, J.E.; Donahue, L.; Li, X.-M.; Hazen, S.L. Measurement of trimethylamine-N-oxide by stable isotope dilution liquid chromatography tandem mass spectrometry. Anal. Biochem. 2014, 455, 35–40. [Google Scholar] [CrossRef] [Green Version]
- Ke, Y.; Li, D.; Zhao, M.; Liu, C.; Liu, J.; Zeng, A.; Shi, X.; Cheng, S.; Pan, B.; Zheng, L.; et al. Gut flora-dependent metabolite Trimethylamine-N-oxide accelerates endothelial cell senescence and vascular aging through oxidative stress. Free Radic. Biol. Med. 2018, 116, 88–100. [Google Scholar] [CrossRef]
- Vogt, N.M.; Romano, K.A.; Darst, B.F.; Engelman, C.D.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Blennow, K.; Zetterberg, H.; Bendlin, B.B.; et al. The gut microbiota-derived metabolite trimethylamine N-oxide is elevated in Alzheimer’s disease. Alzheimer’s Res. Ther. 2018, 10, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrea, L.; Annunziata, G.; Muscogiuri, G.; Di Somma, C.; Laudisio, D.; Maisto, M.; de Alteriis, G.; Tenore, G.C.; Colao, A.; Savastano, S. Trimethylamine-N-oxide (TMAO) as Novel Potential Biomarker of Early Predictors of Metabolic Syndrome. Nutrients 2018, 10, 1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oellgaard, J.; Winther, S.A.; Hansen, T.S.; Rossing, P.; von Scholten, B.J. Trimethylamine N-oxide (TMAO) as a New Potential Therapeutic Target for Insulin Resistance and Cancer. Curr. Pharm. Des. 2017, 23, 3699–3712. [Google Scholar] [CrossRef] [PubMed]
- Ufnal, M.; Zadlo, A.; Ostaszewski, R. TMAO: A small molecule of great expectations. Nutrition 2015, 31, 1317–1323. [Google Scholar] [CrossRef]
- Subramaniam, S.; Fletcher, C. Trimethylamine N-oxide: Breathe new life. Br. J. Pharmacol. 2018, 175, 1344–1353. [Google Scholar] [CrossRef]
- Hero, C.; Svensson, A.-M.; Gidlund, P.; Gudbjornsdottir, S.; Eliasson, B.; Eeg-Olofsson, K. LDL cholesterol is not a good marker of cardiovascular risk in Type 1 diabetes. Diabet. Med. 2016, 33, 316–323. [Google Scholar] [CrossRef]
- Upadhyay, R.K. Emerging risk biomarkers in cardiovascular diseases and disorders. J. Lipids 2015, 2015, 971453. [Google Scholar] [CrossRef]
- Ivanova, E.A.; Myasoedova, V.A.; Melnichenko, A.A.; Grechko, A.V.; Orekhov, A.N. Small Dense Low-Density Lipoprotein as Biomarker for Atherosclerotic Diseases. Oxid. Med. Cell. Longev. 2017, 2017, 1273042. [Google Scholar] [CrossRef]
- Alshahrani, S.M.; Fraser, G.E.; Sabate, J.; Knutsen, R.; Shavlik, D.; Mashchak, A.; Lloren, J.I.; Orlich, M.J. Red and Processed Meat and Mortality in a Low Meat Intake Population. Nutrients 2019, 11, 622. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K.A.; Shea, J.W. Dietary Choline and Betaine and Risk of CVD: A Systematic Review and Meta-Analysis of Prospective Studies. Nutrients 2017, 9, 711. [Google Scholar] [CrossRef] [Green Version]
- Adeva-Andany, M.M.; Calvo-Castro, I.; Fernandez-Fernandez, C.; Donapetry-Garcia, C.; Pedre-Pineiro, A.M. Significance of l-carnitine for human health. IUBMB Life 2017, 69, 578–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ussher, J.R.; Lopaschuk, G.D.; Arduini, A. Gut microbiota metabolism of L-carnitine and cardiovascular risk. Atherosclerosis 2013, 231, 456–461. [Google Scholar] [CrossRef]
- Fukami, K.; Yamagishi, S.; Sakai, K.; Kaida, Y.; Yokoro, M.; Ueda, S.; Wada, Y.; Takeuchi, M.; Shimizu, M.; Yamazaki, H.; et al. Oral L-carnitine supplementation increases trimethylamine-N-oxide but reduces markers of vascular injury in hemodialysis patients. J. Cardiovasc. Pharmacol. 2015, 65, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Olek, R.A.; Samulak, J.J.; Sawicka, A.K.; Hartmane, D.; Grinberga, S.; Pugovics, O.; Lysiak-Szydlowska, W. Increased Trimethylamine N-Oxide Is Not Associated with Oxidative Stress Markers in Healthy Aged Women. Oxid. Med. Cell. Longev. 2019, 2019, 6247169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.Q.; Mitchell, S.C.; Smith, R.L. Dietary precursors of trimethylamine in man: A pilot study. Food Chem. Toxicol. 1999, 37, 515–520. [Google Scholar] [CrossRef]
- Landfald, B.; Valeur, J.; Berstad, A.; Raa, J. Microbial trimethylamine-N-oxide as a disease marker: Something fishy? Microb. Ecol. Health Dis. 2017, 28, 1327309. [Google Scholar] [CrossRef] [Green Version]
- Tong, T.Y.N.; Appleby, P.N.; Bradbury, K.E.; Perez-Cornago, A.; Travis, R.C.; Clarke, R.; Key, T.J. Risks of ischaemic heart disease and stroke in meat eaters, fish eaters, and vegetarians over 18 years of follow-up: Results from the prospective EPIC-Oxford study. BMJ 2019, 366, l4897. [Google Scholar] [CrossRef] [Green Version]
- Yasukawa, T.; Kang, D. An overview of mammalian mitochondrial DNA replication mechanisms. J. Biochem. 2018, 164, 183–193. [Google Scholar] [CrossRef]
- Janssen, B.G.; Byun, H.M.; Cox, B.; Gyselaers, W.; Izzi, B.; Baccarelli, A.A.; Nawrot, T.S. Variation of DNA methylation in candidate age-related targets on the mitochondrial-telomere axis in cord blood and placenta. Placenta 2014, 35, 665–672. [Google Scholar] [CrossRef] [Green Version]
- Mishra, M.; Kowluru, R.A. Epigenetic Modification of Mitochondrial DNA in the Development of Diabetic Retinopathy. Invest. Ophthalmol. Vis. Sci. 2015, 56, 5133–5142. [Google Scholar] [CrossRef]
- Feng, S.; Xiong, L.; Ji, Z.; Cheng, W.; Yang, H. Correlation between increased ND2 expression and demethylated displacement loop of mtDNA in colorectal cancer. Mol. Med. Rep. 2012, 6, 125–130. [Google Scholar] [PubMed] [Green Version]
- Byun, H.-M.; Colicino, E.; Trevisi, L.; Fan, T.; Christiani, D.C.; Baccarelli, A.A. Effects of Air Pollution and Blood Mitochondrial DNA Methylation on Markers of Heart Rate Variability. J. Am. Heart Assoc. 2016, 5, e003218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Wijst, M.G.P.; van Tilburg, A.Y.; Ruiters, M.H.J.; Rots, M.G. Experimental mitochondria-targeted DNA methylation identifies GpC methylation, not CpG methylation, as potential regulator of mitochondrial gene expression. Sci. Rep. 2017, 7, 177. [Google Scholar] [CrossRef] [Green Version]
- Pecina, P.; Houstkova, H.; Hansikova, H.; Zeman, J.; Houstek, J. Genetic defects of cytochrome c oxidase assembly. Physiol. Res. 2004, 53, S213–S223. [Google Scholar] [PubMed]
- Lorente, L.; Martin, M.M.; Lopez-Gallardo, E.; Iceta, R.; Blanquer, J.; Sole-Violan, J.; Labarta, L.; Diaz, C.; Jimenez, A.; Montoya, J.; et al. Higher platelet cytochrome oxidase specific activity in surviving than in non-surviving septic patients. Crit. Care 2014, 18, R136. [Google Scholar] [CrossRef] [Green Version]
- Garai, B.; Chatterjee, S.; Mondal, S.; Mondal, T. Effect of exercise on platelet variables: An overview. Int. J. Phys. Educ. Sport. Health 2017, 4, 506–510. [Google Scholar]
- Creighton, B.C.; Kupchak, B.R.; Aristizabal, J.C.; Flanagan, S.D.; Dunn-Lewis, C.; Volk, B.M.; Comstock, B.A.; Volek, J.S.; Hooper, D.R.; Szivak, T.K.; et al. Influence of training on markers of platelet activation in response to a bout of heavy resistance exercise. Eur. J. Appl. Physiol. 2013, 113, 2203–2209. [Google Scholar] [CrossRef]
- Mohammadi, M.; Hajhossein Talasaz, A.; Alidoosti, M. Preventive effect of l-carnitine and its derivatives on endothelial dysfunction and platelet aggregation. Clin. Nutr. ESPEN 2016, 15, 1–10. [Google Scholar] [CrossRef]
- Chang, J.-C.; Kou, S.-J.; Lin, W.-T.; Liu, C.-S. Regulatory role of mitochondria in oxidative stress and atherosclerosis. World J. Cardiol. 2010, 2, 150–159. [Google Scholar] [CrossRef]
- Wang, J.; Lin, F.; Guo, L.-L.; Xiong, X.-J.; Fan, X. Cardiovascular Disease, Mitochondria, and Traditional Chinese Medicine. Evid. Based Complement. Alternat. Med. 2015, 2015, 143145. [Google Scholar] [CrossRef]
- Ashar, F.N.; Zhang, Y.; Longchamps, R.J.; Lane, J.; Moes, A.; Grove, M.L.; Mychaleckyj, J.C.; Taylor, K.D.; Coresh, J.; Rotter, J.I.; et al. Association of Mitochondrial DNA Copy Number With Cardiovascular Disease. JAMA Cardiol. 2017, 2, 1247–1255. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.S.G.; Simoes, R.F.; Couto, R.; Oliveira, P.J. Targeting Mitochondria in Cardiovascular Diseases. Curr. Pharm. Des. 2016, 22, 5698–5717. [Google Scholar] [CrossRef] [PubMed]
- Huc, T.; Drapala, A.; Gawrys, M.; Konop, M.; Bielinska, K.; Zaorska, E.; Samborowska, E.; Wyczalkowska-Tomasik, A.; Paczek, L.; Dadlez, M.; et al. Chronic, low-dose TMAO treatment reduces diastolic dysfunction and heart fibrosis in hypertensive rats. Am. J. Physiol. Heart Circ. Physiol. 2018, 315, H1805–H1820. [Google Scholar] [CrossRef]
- Cho, C.E.; Taesuwan, S.; Malysheva, O.V.; Bender, E.; Tulchinsky, N.F.; Yan, J.; Sutter, J.L.; Caudill, M.A. Trimethylamine-N-oxide (TMAO) response to animal source foods varies among healthy young men and is influenced by their gut microbiota composition: A randomized controlled trial. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef]
- Ventura-Clapier, R.; Moulin, M.; Piquereau, J.; Lemaire, C.; Mericskay, M.; Veksler, V.; Garnier, A. Mitochondria: A central target for sex differences in pathologies. Clin. Sci. (Lond.) 2017, 131, 803–822. [Google Scholar] [CrossRef] [PubMed]
- Scicchitano, P.; Cameli, M.; Maiello, M.; Modesti, P.A.; Muiesan, M.L.; Novo, S.; Palmiero, P.; Saba, P.S.; Pedrinelli, R.; Ciccone, M.M. Nutraceuticals and dyslipidaemia: Beyond the common therapeutics. J. Funct. Foods 2014, 6, 11–32. [Google Scholar] [CrossRef]
- Peluso, I.; Palmery, M.; Drummen, G. Biomarkers of Oxidative Stress in Experimental Models and Human Studies with Nutraceuticals: Measurement, Interpretation, and Significance 2017. Oxid. Med. Cell. Longev. 2017, 2017, 3457917. [Google Scholar] [CrossRef]
- Bell, K.E.; Snijders, T.; Zulyniak, M.; Kumbhare, D.; Parise, G.; Chabowski, A.; Phillips, S.M. A whey protein-based multi-ingredient nutritional supplement stimulates gains in lean body mass and strength in healthy older men: A randomized controlled trial. PLoS ONE 2017, 12, e0181387. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.-H.; Park, K.S.; Ahn, S.; Ku, E.J.; Jung, K.Y.; Kim, Y.J.; Kim, K.M.; Moon, J.H.; Choi, S.H.; Park, K.S.; et al. Comparison of Abdominal Visceral Adipose Tissue Area Measured by Computed Tomography with That Estimated by Bioelectrical Impedance Analysis Method in Korean Subjects. Nutrients 2015, 7, 10513–10524. [Google Scholar] [CrossRef] [Green Version]
- Jaworska, K.; Huc, T.; Samborowska, E.; Dobrowolski, L.; Bielinska, K.; Gawlak, M.; Ufnal, M. Hypertension in rats is associated with an increased permeability of the colon to TMA, a gut bacteria metabolite. PLoS ONE 2017, 12, e0189310. [Google Scholar] [CrossRef] [Green Version]
- Knopfholz, J.; Disserol, C.C.D.; Pierin, A.J.; Schirr, F.L.; Streisky, L.; Takito, L.L.; Massucheto Ledesma, P.; Faria-Neto, J.R.; Olandoski, M.; da Cunha, C.L.P.; et al. Validation of the friedewald formula in patients with metabolic syndrome. Cholesterol 2014, 2014, 261878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Du, Q.; Chen, L.; Fu, G.; Li, S.; Fu, L.; Zhang, X.; Ma, C.; Bin, C. CpG methylation patterns of human mitochondrial DNA. Sci. Rep. 2016, 6, 23421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, Q.; Fan, Z.; Xie, R.; Wang, Z.; Lu, Y. Platelet mitochondrial dysfunction and the correlation with human diseases. Biochem. Soc. Trans. 2017, 45, 1213–1223. [Google Scholar] [CrossRef] [PubMed]
- Zharikov, S.; Shiva, S. Platelet mitochondrial function: From regulation of thrombosis to biomarker of disease. Biochem. Soc. Trans. 2013, 41, 118–123. [Google Scholar] [CrossRef]







| Mean ± SD | Range | Shapiro–Wilk (p Value) | |
|---|---|---|---|
| Height (cm) | 159.9 ± 5.2 | 151–175 | 0.241 |
| Age (years) | 67.3 ± 2.7 | 62–72 | 0.284 |
| Basal Metabolic Rate | 1342 ± 98 | 1183–1559 | 0.500 |
| Obesity Degree | 130.3 ± 20.7 | 90.8–165.0 | 0.075 |
| Visceral Fat Area (cm2) | 107.3 ± 36.4 | 40.5–170.8 | 0.268 |
| Weight (kg) | 71.9 ± 13.1 | 50.6–93.3 | 0.104 |
| Skeletal Muscle Mass (kg) | 24.6 ± 2.7 | 20.2–30.0 | 0.434 |
| Body Fat Mass (kg) | 26.9 ± 10.5 | 9.9–46.8 | 0.271 |
| Body Fat (%) | 36.1 ± 8.5 | 19.5–51.5 | 0.555 |
| Body Mass Index | 28.0 ± 4.4 | 19.5–35.5 | 0.076 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bordoni, L.; Sawicka, A.K.; Szarmach, A.; Winklewski, P.J.; Olek, R.A.; Gabbianelli, R. A Pilot Study on the Effects of l-Carnitine and Trimethylamine-N-Oxide on Platelet Mitochondrial DNA Methylation and CVD Biomarkers in Aged Women. Int. J. Mol. Sci. 2020, 21, 1047. https://doi.org/10.3390/ijms21031047
Bordoni L, Sawicka AK, Szarmach A, Winklewski PJ, Olek RA, Gabbianelli R. A Pilot Study on the Effects of l-Carnitine and Trimethylamine-N-Oxide on Platelet Mitochondrial DNA Methylation and CVD Biomarkers in Aged Women. International Journal of Molecular Sciences. 2020; 21(3):1047. https://doi.org/10.3390/ijms21031047
Chicago/Turabian StyleBordoni, Laura, Angelika K. Sawicka, Arkadiusz Szarmach, Pawel J. Winklewski, Robert A. Olek, and Rosita Gabbianelli. 2020. "A Pilot Study on the Effects of l-Carnitine and Trimethylamine-N-Oxide on Platelet Mitochondrial DNA Methylation and CVD Biomarkers in Aged Women" International Journal of Molecular Sciences 21, no. 3: 1047. https://doi.org/10.3390/ijms21031047
APA StyleBordoni, L., Sawicka, A. K., Szarmach, A., Winklewski, P. J., Olek, R. A., & Gabbianelli, R. (2020). A Pilot Study on the Effects of l-Carnitine and Trimethylamine-N-Oxide on Platelet Mitochondrial DNA Methylation and CVD Biomarkers in Aged Women. International Journal of Molecular Sciences, 21(3), 1047. https://doi.org/10.3390/ijms21031047